













| General Information: |
|
| Name(s) found: |
MUS81_ORYSJ
[Swiss-Prot]
|
| Description(s) found:
Found 8 descriptions. SHOW ALL |
|
| Organism: | Oryza sativa Japonica Group |
| Length: | 660 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAPEARQLKV HLRENEAVAQ CVLEKWRSME EKPGGLKENL AHTLYKSYRN VCAAKEPIRS 60
61 LKDLYQIKGV GKWVIRQLKG SFPESSPDLS PPESNAAGEK GKKAGGSKRY VPQKNSAAYA 120
121 ILITLHRETI NGKSHMKKQE LIDATEASGL SRSAIGPDKS KAKPGAFASS QKDWYTGWSC 180
181 MKTLTSKGLV AKSGNPAKYM ITEEGKSTAL ECLSRSGLDD HAAPLVINSA PDTSNASHKL 240
241 NNICMTSFVE TSSGPSRAIG RPKTSIANPA TKTSPEVTYL TSQESLNYNS DVRTAENCAE 300
301 EIILSDSDSE ELYTENYPLI GSEEFTERVA PPILNASNSG KTTTNYRFSD CSASISPRSS 360
361 EGTFEMQSSS TMGIAEFNML DNDTVCMDNS ILAMPPRRSS KNFLEDYEVV LILDDRENFG 420
421 GRSRKTVDNI HSQFRVPVEI KHLPVGDGIW IARDRKLHTE YVLDFIVERK NVADLCSSIT 480
481 DNRYKDQKLR LKKCGLRKLI YLVEGDPNPL DTSERIKTAC FTTEILEGFD VQRTPGYAET 540
541 VRTYGNLTHS ITEYYSTHFS TGANTSQVCL TYDEFTKKCD DLKKITVSDV FALQLMQVPQ 600
601 VTEEAALAVI GLYPTLFSLA KAYSMLDGDT HAQEKMLKNK STLINAGASR NIFKLVWAEG 660
661 |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
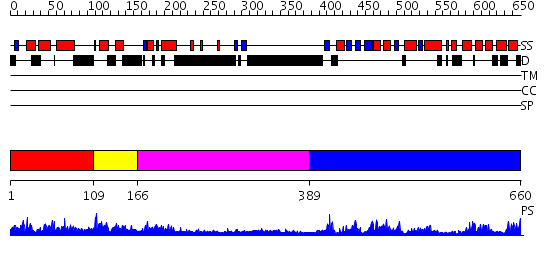
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..108] | 2.019996 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [109..165] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [166..388] | 13.39794 | Crystal structure of Pyrococcus furiosus Hef helicase domain | |
| 4 | View Details | [389..660] | 34.39794 | XPF from Aeropyrum pernix, complex with DNA |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)