













| General Information: |
|
| Name(s) found: |
HGNC:13579|Ensembl:ENSG00000159086|HPRD:10761,HsxXG003541,C05,HsCD00302978,470535.0,pANT7_cGST,FUSION,NM_016631,94104.0,GCFC1,BM020|C21orf66|FLJ90561|FSAP105|GCFC,GC-rich
[120926-122011-mus-fusion.fasta]
|
| Description(s) found: |
|
| Organism: | SPECIES UNKNOWN |
| Length: | 917 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MFRKARRVNV RKRNDSEEEE RERDEEQEPP PLLPPPGTGE EAGPGGGDRA PGGESLLGPG 60
61 PSPPSALTPG LGAEAGGGFP GGAEPGNGLK PRKRPRENKE VPRASLLSFQ DEEEENEEVF 120
121 KVKKSSYSKK IVKLLKKEYK EDLEKSKIKT ELNSSAESEQ PLDKTGHVKD TNQEDGVIIS 180
181 EHGEDEMDME SEKEEEKPKT GGAFSNALSS LNVLRPGEIP DAAFIHAARK KRQMARELGD 240
241 FTPHDNEPGK GRLVREDEND ASDDEDDDEK RRIVFSVKEK SQRQKIAEEI GIEGSDDDAL 300
301 VTGEQDEELS RWEQEQIRKG INIPQVQASQ PAEVNMYYQN TYQTMPYGSS YGIPYSYTAY 360
361 GSSDAKSQKT DNTVPFKTPS NEMTPVTIDL VKKQLKDRLD SMKELHKTNR QQHEKHLQSR 420
421 VDSTRAIERL EGSSGGIGER YKFLQEMRGY VQDLLECFSE KVPLINELES AIHQLYKQRA 480
481 SRLVQRRQDD IKDESSEFSS HSNKALMAPN LDSFGRDRAL YQEHAKRRIA EREARRTRRR 540
541 QAREQTGKMA DHLEGLSSDD EETSTDITNF NLEKDRISKE SGKVFEDVLE SFYSIDCIKS 600
601 QFEAWRSKYY TSYKDAYIGL CLPKLFNPLI RLQLLTWTPL EAKCRDFENM LWFESLLFYG 660
661 CEEREQEKDD VDVALLPTIV EKVILPKLTV IAENMWDPFS TTQTSRMVGI TLKLINGYPS 720
721 VVNAENKNTQ VYLKALLLRM RRTLDDDVFM PLYPKNVLEN KNSGPYLFFQ RQFWSSVKLL 780
781 GNFLQWYGIF SNKTLQELSI DGLLNRYILM AFQNSEYGDD SIKKAQNVIN CFPKQWFMNL 840
841 KGERTISQLE NFCRYLVHLA DTIYRNSIGC SDVEKRNARE NIKQIVKLLA SVRALDHAMS 900
901 VASDHNVKEF KSLIEGK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
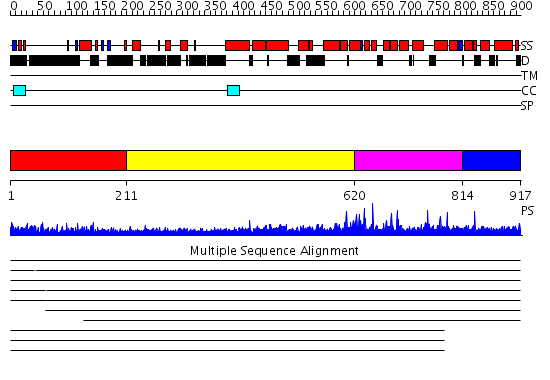
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..210] | 5.522879 | No description for 1y0fA was found. | |
| 2 | View Details | [211..619] | 6.522879 | Heavy meromyosin subfragment | |
| 3 | View Details | [620..813] | 1.043982 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [814..917] | 1.035958 | View MSA. Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)