













| General Information: |
|
| Name(s) found: |
gi|24374363
[NCBI NR]
gi|24348920 [NCBI NR] |
| Description(s) found:
Found 5 descriptions. SHOW ALL |
|
| Organism: | Shewanella oneidensis MR-1 |
| Length: | 705 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: |
2'-deoxyribonucleotide biosynthetic process
[ISS |
| Molecular Function: |
ribonucleoside-triphosphate reductase activity
[ISS |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPVVIKRDGC RTPFDETRIR DAVIAAANSV GIEDTDYAAT VAACVSARVA NMAEVEIHHL 60
61 QDVVENLLME GPYKSLARVY IEYRHDRDIC RDATSKLNLE IRGLVEQSNA ALLNENANKD 120
121 SKVIPTQRDL LAGIVAKHYA KSHILPKDVV AAHEMGEIHY HDLDYSPFFP MFNCMLIDLA 180
181 GMLTHGFKMG NAEIETPKSI STATAVTAQI IAQVASHIYG GTTINRIDEV LAPFVAKSYD 240
241 KHYQVALNWQ IKDAKAYATA QTEKECHDAF QSLEYEVNTL HTANGQTPFV TFGFGLGTSW 300
301 ESRLIQQSML KVRMAGLGKN RKTAVFPKLV FAIRDGINHK VGDCNYDVKQ LALKCATMRM 360
361 YPDILNYEQV EKVTGSFKTP MGCRSFLGSY KENGELVHEG RNNLGVVSLN LPRVALEAKG 420
421 DEAEFYRILD ERLLLARRAL DTRIERLNGV KARVAPILYM EGACGVRLRA DDDIAQIFKN 480
481 GRASISLGYI GLHETINALY GTETHVFDNA QLREKAVAIV AYLKAATDAW KAETGYGFSL 540
541 YSTPSESLCS RFCKLDARQF GVVTGVTDKG YYTNSFHLDV EKRVNPYDKI DFEQPYPAIA 600
601 NGGFICYGEY PNMQHNIEAL ENVWDYSYSR VPYYGTNTPI DECYDCGFTG EFSCTSKGFT 660
661 CPKCGNHEPS RVSVTRRVCG YLGSPDARPF NHGKQEEVKR RIKHL |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
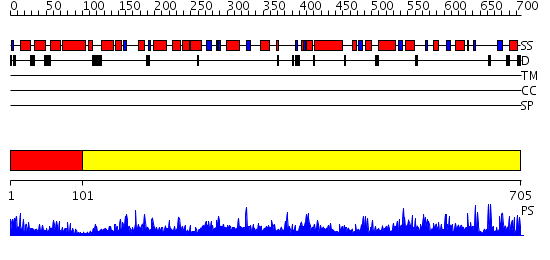
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..100] | 3.221849 | R1 subunit of ribonucleotide reductase, N-terminal domain; R1 subunit of ribonucleotide reductase, C-terminal domain | |
| 2 | View Details | [101..705] | 1000.0 | Class III anaerobic ribonucleotide reductase NRDD subunit |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||||||||
| 1 | No functions predicted. | |||||||||||||||||||||||||||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)