













| General Information: |
|
| Name(s) found: |
ZRG8_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1076 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRSFIKAHKK STSFDESPKR HSNFSGNTNN SSQRSSDDSL DFLPSTPSQM NYDSIPPPAK 60
61 HSPGFESFHR LANKTSKLFK KTSNSNLNSH LASTPTTSTN QTTSNSFVLQ NPPTKNTGPP 120
121 PPLPPPLFPS SSTSSFSRHD NESEYTAYKK TSPAKDFNRT TDSLPAIKGT ITHSWGDSKV 180
181 ESHVIILNDP ASPASNTSEA TSSKQFKTPI IGNENLTSTT SPSNLEPAIR ILNKNKGKQQ 240
241 ENIDDAEDGS SKKEHHVYKA LALAKNRNRQ ARIHSHDDII NLGKASQMDM SLLAAAFSGN 300
301 STTTINNDQS SNEQTDEKIL DIERVTTTST LTSSETTSPI NKSPCFYSQT LSLSPKIRHG 360
361 DLQSSPSKVN KNDSQNETLN KKKVRISLNR KEEEKVYSLN NNSDEYSVNE KETHKANDCN 420
421 DESSENGDGD NDHDDDYDDD DDDDDDDDES EFSFEYAGIN VRTSSVKYYS KPEPAANVYI 480
481 DDLYEDENFD DDMNCIEDDE SGNEGNEICG LSTRFEETSL KSNKVKKFND LFNLSDDDEE 540
541 EDGKDNSNNG DENESDNLYQ KRLENGKETF NGNHGGHHDD ASLGETVDNK EQFLINDNVK 600
601 KPIQKYNDLF DLSDEDDNDD KEMSEAESYM FSDEAPSIES GPANAKSTRG IYSQSNKNII 660
661 RDGKPNYSFS LKRNNSDDET EHTSAIKASL TGTTGSTKPT VKSFSDIFNV DDSASDAESD 720
721 SGTGGNNSNG LVSNDSERQV SLQSSLYETK SESHPPNHPH SQILQTPAKI VITPSVSDAQ 780
781 SQALAITDDD GEDDDDDTSS ILRTPFQLID SSHSQQPHYA SPQYTAVLNS PPLPPPARSQ 840
841 SLKYHDLNCD LDSEVPRPMS NLFFIDEAEE DEYNQKSKFF DFDHYDIDEI NGIPEDFNFS 900
901 DSERDDLNRR TLKSPLRGGS KNREVSPFSS VSSSFRSTHS FNGKLTINQG AKELAPMKNK 960
961 IELTNKTVTF FNSNNWNTYD CNSLSRKTSS QMRDSKYQNH NVGQNVEPSS VLSPQHQISN1020
1021 GLDGKCNDNY VISPNLPTTI TPTNSFTKPT PEFSNDYSLS PIQETPSSVQ SSPKRA |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
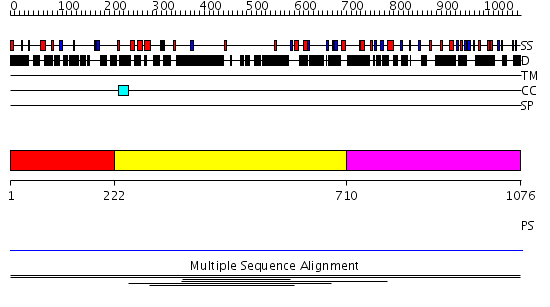
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..221] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [222..709] | 1.004997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [710..1076] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [812..1076] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)