













| General Information: |
|
| Name(s) found: |
YJU6_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 758 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTPKESGKPI SCAMKKLKGK RSKILVLSRD AGTNELKPTK GRAHRACIAC RKRKVRCSGN 60
61 IPCRLCQTNS YECKYDRPPR NSSVFDREVS DDSSLYAQRA SHEREDSKGP ISSIDYKKVV 120
121 ETIFPPETLR QILASSSFNS QNFLDTIKTC LLQGQLNVNQ VIRQSLPKDT PWHMQTSVPL 180
181 PPREIALKFI QKTWDCACVL FRFYHRPTII SILDSIYEAE KHGKQYTPEQ VKTQPLIYSV 240
241 LAVGALFSKE DLSKDSKATR EFYTDEGYRY FLEAKNSLDF SNITDIYSIQ AIFMMTIFLQ 300
301 CSANLKACYS FIGIALRAAL KEGLHRRSSI VGPTPIQDET KKRLFWSVYK LDLYMNCILG 360
361 FPSGIDESDI DQEFPLDVDD ENISTIGIKF QDWRTISSCG MNNKHTKLIL IMSRIYKLMY 420
421 SLRRKPLEED SRTQIVSLND QLDNWYAQLP DILKVDTIRY RQTQPPLTVS ANDTSSPYTK 480
481 PKKLLYLDFL LSKIVLYKPF YHYISIDPLD IPEFQFQIHM AENCIEVAKK VIQLSYEMIT 540
541 QNLLSGSYWF SIHTIFFSVA CLKFYVYQTE KGLIRNGKVD SDIHNATQLG SEILSLLKGA 600
601 SNASKRTFEV LNQLFKEFNE KTSVLSEQLL NIVKLQRQES SGALVPQLQT NNNFTKCQGE 660
661 LHHGQQHHQT PATSLRSILN LPQGEADLKF QNTNNESHTT TAAQEEYLDK LLAEFEEFDY 720
721 SINRVLPDVI DFSALIGQDS SANNQIFSSE FSSDPTVN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
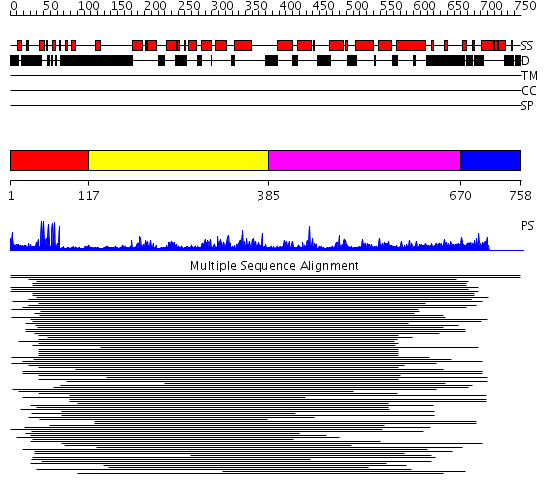
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..116] | 39.917458 | PUT3 | |
| 2 | View Details | [117..384] | 2.148988 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [385..669] | 1.113987 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [670..758] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [622..758] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 |
|
||||||
| 2 | No functions predicted. | ||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. | ||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.56 |
Source: Reynolds et al. (2008)