













| General Information: |
|
| Name(s) found: |
YJH0_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQAVERRPSL LFDEYQNSVT KPNETKNKEA RVLSENDGDV SPSVLKQKEI SVDDMDMISL 60
61 PTEFDRQMVL GSPMFFDLED EENKIDPLPS VSHHYGNGES DSFVSSYTPS NLKTGEETKD 120
121 LFINPFELVS QMRKRYIAAS KQDGISNIKN DTEKWFLYPK PLPKFWRFED DKRFQDPSDS 180
181 DLNDDGDSTG TGAATPHRHG YYYPSYFTDH YYYYTKSGLK GKGNIKVPYT GEYFDLEDYK 240
241 KQYIYHLSNQ ENTQNPLSPY SSKEESLEEE FLTDVPTFQE FRDDFAYIIE LIQSHKFNEV 300
301 SRKRLSYLLD KFELFQYLNS KKEILANKNV PYRDFYNSRK VDRDLSLSGC ISQRQLSEYI 360
361 WEKINLEPER IVYQDPETSR KLSLRDIFQF GCSSNDQPIA IGLKLIDDEF LDWYRNIYLI 420
421 DYHLTPNKVA KLVGKEMRFY LLAKVFLEFD NFIEGEYLAE IFIKYVIHIL EKSKYQLAQV 480
481 SVNFQFYSSG EDWYKKFSQW LLRWKLVSYN IRWNIQIARI FPKLFKENVV SNFQEFLDLI 540
541 FNPLFTLEKE QLPIDSSVNT DIIGLQFFLS NVCSMDLVIK ESDEYYWKEF TDMNCKPKFW 600
601 TAQGDNPTVA HYMYYIYKSL AKVNFLRSQN LQNTITLRNY CSPLSSRTSQ FGVDLYFTDQ 660
661 VESLVCNLLL CNGGLLQVEP LWDTATMIQY LFYLFQIPIL AAPLSSVSLL NSQKSTFLKN 720
721 KNVLLEHDYL KDQETAKINP SRDITVGEQR SYETNPFMKM FKMGLKISLS SKSILYNSSY 780
781 TLEPLIEEYS VAASIYLLNP TDLCELSRTS VLSSGYEGWY KAHWIGVGVK KAPYFEENVG 840
841 GIDNWYDTAK DTSIKHNVPM IRRRYRKETL DQEWNFVRDH FGVINSIW |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
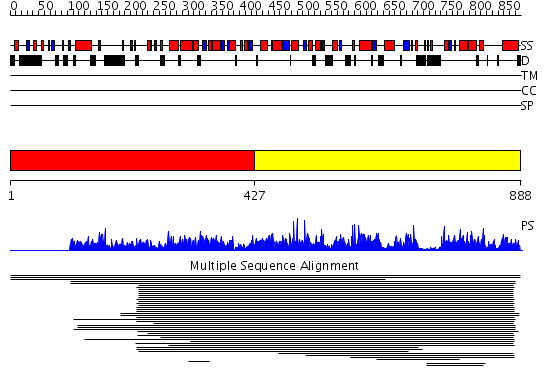
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..426] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [427..888] | 14.57 | No description for 1m7mA was found. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)