













| General Information: |
|
| Name(s) found: |
YAK1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 807 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNSSNNNDSS SSNSNMNNSL SPTLVTHSDA SMGSGRASPD NSHMGRGIWN PSYVNQGSQR 60
61 SPQQQHQNHH QQQQQQQQQQ QQNSQFCFVN PWNEEKVTNS QQNLVYPPQY DDLNSNESLD 120
121 AYRRRKSSLV VPPARAPAPN PFQYDSYPAY TSSNTSLAGN SSGQYPSGYQ QQQQQVYQQG 180
181 AIHPSQFGSR FVPSLYDRQD FQRRQSLAAT NYSSNFSSLN SNTNQGTNSI PVMSPYRRLS 240
241 AYPPSTSPPL QPPFKQLRRD EVQGQKLSIP QMQLCNSKND LQPVLNATPK FRRASLNSKT 300
301 ISPLVSVTKS LITTYSLCSP EFTYQTSKNP KRVLTKPSEG KCNNGFDNIN SDYILYVNDV 360
361 LGVEQNRKYL VLDILGQGTF GQVVKCQNLL TKEILAVKVV KSRTEYLTQS ITEAKILELL 420
421 NQKIDPTNKH HFLRMYDSFV HKNHLCLVFE LLSNNLYELL KQNKFHGLSI QLIRTFTTQI 480
481 LDSLCVLKES KLIHCDLKPE NILLCAPDKP ELKIIDFGSS CEEARTVYTY IQSRFYRAPE 540
541 IILGIPYSTS IDMWSLGCIV AELFLGIPIF PGASEYNQLT RIIDTLGYPP SWMIDMGKNS 600
601 GKFMKKLAPE ESSSSTQKHR MKTIEEFCRE YNIVEKPSKQ YFKWRKLPDI IRNYRYPKSI 660
661 QNSQELIDQE MQNRECLIHF LGGVLNLNPL ERWTPQQAML HPFITKQEFT GEWFPPGSSL 720
721 PGPSEKHDDA KGQQSEYGSA NDSSNNAGHN YVYNPSSATG GADSVDIGAI SKRKENTSGD 780
781 ISNNFAVTHS VQEGPTSAFN KLHIVEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
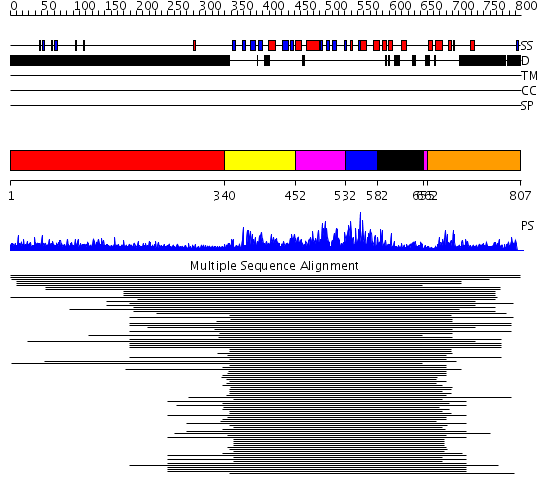
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..339] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [340..451] | 434.0 | Cyclin-dependent PK, CDK2 | |
| 3 | View Details | [452..531] [655..661] |
434.0 | Cyclin-dependent PK, CDK2 | |
| 4 | View Details | [532..581] | 434.0 | Cyclin-dependent PK, CDK2 | |
| 5 | View Details | [582..654] | 434.0 | Cyclin-dependent PK, CDK2 | |
| 6 | View Details | [662..807] | 10.52 | Glycogen synthase kinase-3 beta (Gsk3b) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)