













| General Information: |
|
| Name(s) found: |
VID22_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 901 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MRAMDTQVQS AERGLVLPPM NSTVSSATAA TTATNTDTDT DGDRDEERES LAEDGSEWVP 60
61 AYMLTRDRSR YLGHFLGVDK MLEAVKCKYC GVIIRRQGNS ISMAEASQTH LWSTHKIDPN 120
121 ANYYSGWTGV EAGSTFMVRP PLKNHQGGSA TTNSIANLLE IDEDFLKRTR EKEMALPLVQ 180
181 SLAIIIASEN LPLSFVDNTA VRLLINQNAN SLSFIDHDLI LNAIRSIAYN LDRIIQRTAL 240
241 RNNSDLSLII DKNYLLMDPT DRSNQLSNRL KNQLFEMQKI NFFSLSHSVW NNTISILSIQ 300
301 YYDDFHSQVK TLPLIIQNLH EYNNDPKLSI PAQLLKISQE LPGLQNTVIS ITLPRSQIVD 360
361 LLNVMDSQPF FPNTYTNAKN YYHNCIISII NSAILPLFGT PKSADITHPR QSSFSKEPLT 420
421 LLDSLIDLSN IDISNSIFSR INSFLDDLQS NSWQLDKFRS LCEKFGFEFV CSKFDLSRYS 480
481 TATVSLQTFL NLRPIIEEYQ SSIQIEKFNE IDFQIIDYLL ITLNSINRIL KFFTSSKSLN 540
541 FTYVLFAIMS IEKHLLSTLG SLQFQRLIAP FETFLSKIQE FKTILFSDDM NLLAMFLCPA 600
601 ILFEREVLEY SFHTISLSEI VDKLSTSIFS LLKRFLNLHT IGNVNNSHNT SNHSNMNIHT 660
661 DNQTNNINNR SGNNSDNNDN EHDNDNDNHS NSNTPASRID IDPTGGENSV LPEQQPQNSN 720
721 NNLSFGSLSD THHLSDSTIS KEIDSIFLQI IQEDLYDYLS TVNSIVPISY RSYCEQSNFI 780
781 RDSGRFKKRI ITEDSIIGEL EQPMNFIEEL LDIHVPVCNA FWSQYLDNDA GPIIRILFKI 840
841 MQCQSSSSIR GEYSFLNDFI PRVHPDLTQE IIKIKLFNDQ FVASKVDYDL DTLQTASQYL 900
901 P |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
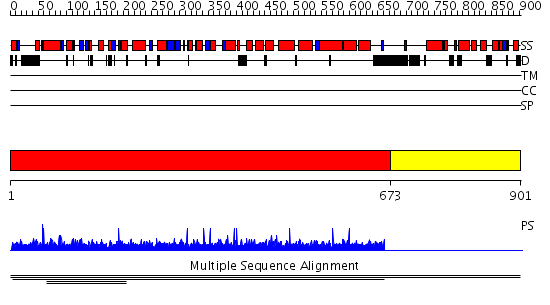
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..672] | 1.002001 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [673..901] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [208..412] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [413..647] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [648..718] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [719..901] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.82 |
Source: Reynolds et al. (2008)