













| General Information: |
|
| Name(s) found: |
UTP5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 643 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSPVLQSAY DPSGQYLCYV TVALDKQRVG VQPTQRATSS GVDTVWNENF LYLEDSKLKV 60
61 TCLKWVNLAS SDTVAIILGM NNGEIWLYSV LANEVTYKFT TGNSYEIKDI DLMGNQLWCI 120
121 DSSDAFYQFD LLQFKLLQHF RINNCVQLNK LTIVPAGDSV AQLLVASHSI SLIDIEEKKV 180
181 VMTFPGHVSP VSTLQVITNE FFISGAEGDR FLNVYDIHSG MTKCVLVAES DIKELSHSGQ 240
241 ADSIAVTTED GSLEIFVDPL VSSSTKKRGN KSKKSSKKIQ IVSKDGRKVP IYNAFINKDL 300
301 LNVSWLQNAT MPYFKNLQWR EIPNEYTVEI SLNWNNKNKS ADRDLHGKDL ASATNYVEGN 360
361 ARVTSGDNFK HVDDAIKSWE RELTSLEQEQ AKPPQANELL TETFGDKLES STVARISGKK 420
421 TNLKGSNLKT ATTTGTVTVI LSQALQSNDH SLLETVLNNR DERVIRDTIF RLKPALAVIL 480
481 LERLAERIAR QTHRQGPLNV WVKWCLIIHG GYLVSIPNLM STLSSLHSTL KRRSDLLPRL 540
541 LALDARLDCT INKFKTLNYE AGDIHSSEPV VEEDEDDVEY NEELDDAGLI EDGEESYGSE 600
601 EEEEGDSDNE EEQKHTSSKQ DGRLETEQSD GEEEAGYSDV EME |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
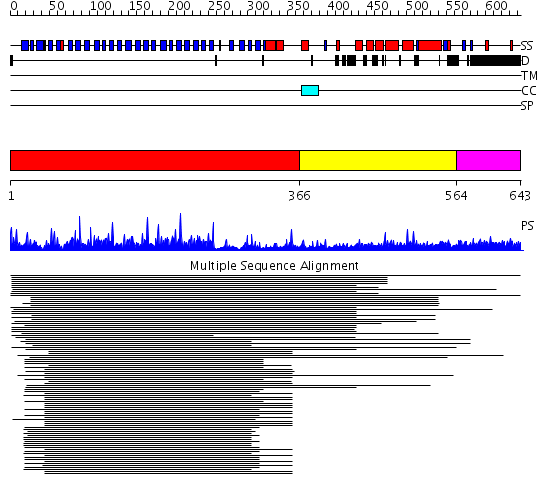
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..365] | 84.450636 | Tup1, C-terminal domain | |
| 2 | View Details | [366..563] | 3.42499 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [564..643] | 1.026992 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [552..643] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.75 |
Source: Reynolds et al. (2008)