













| General Information: |
|
| Name(s) found: |
UTP14_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 899 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAKKKSKSRS KSSRRVLDAL QLAEREINGE FDNSSDNDKR HDARRNGTVV NLLKRSKGDT 60
61 NSDEDDIDSE SFEDEELNSD EALGSDDDYD ILNSKFSQTI RDKKENANYQ EEEDEGGYTS 120
121 IDEEDLMPLS QVWDMDEKTA QSNGNDDEDA SPQLKLQDTD ISSESSSSEE SESESEDDEE 180
181 EEDPFDEISE DEEDIELNTI TSKLIDETKS KAPKRLDTYG SGEANEYVLP SANAASGASG 240
241 KLSLTDMMNV IDDRQVIENA NLLKGKSSTY EVPLPQRIQQ RHDRKAAYEI SRQEVSKWND 300
301 IVQQNRRADH LIFPLNKPTE HNHASAFTRT QDVPQTELQE KVDQVLQESN LANPEKDSKF 360
361 EELSTAKMTP EEMRKRTTEM RLMRELMFRE ERKARRLKKI KSKTYRKIKK KELMKNRELA 420
421 AVSSDEDNED HDIARAKERM TLKHKTNSKW AKDMIKHGMT NDAETREEME EMLRQGERLK 480
481 AKMLDRNSDD EEDGRVQTLS DVENEEKENI DSEALKSKLG KTGVMNMAFM KNGEAREREA 540
541 NKETLRQLRA VENGDDIKLF ESDEEETNGE NIQINKGRRI YTPGSLESNK DMNELNDHTR 600
601 KENKVDESRS LENRLRAKNS GQSKNARTNA EGAIIVEEES DGEPLQDGQN NQQDEEAKDV 660
661 NPWLANESDE EHTVKKQSSK VNVIDKDSSK NVKAMNKMEK AELKQKKKKK GKSNDDEDLL 720
721 LTADDSTRLK IVDPYGGSDD EQGDNVFMFK QQDVIAEAFA GDDVVAEFQE EKKRVIDDED 780
781 DKEVDTTLPG WGEWAGAGSK PKNKKRKFIK KVKGVVNKDK RRDKNLQNVI INEKVNKKNL 840
841 KYQSSAVPFP FENREQYERS LRMPIGQEWT SRASHQELIK PRIMTKPGQV IDPLKAPFK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
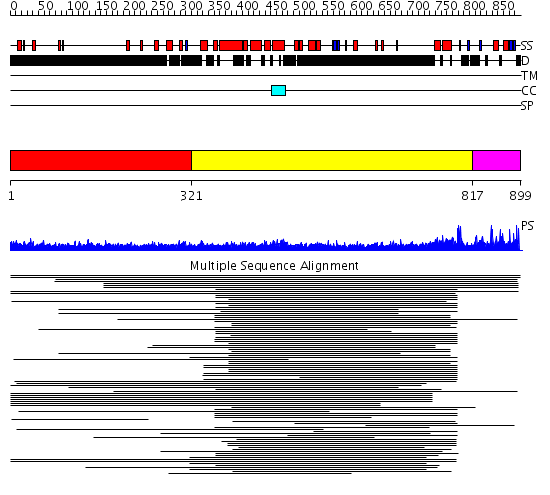
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..320] | 6.522879 | Heavy meromyosin subfragment | |
| 2 | View Details | [321..816] | 14.287996 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [817..899] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [643..762] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [763..899] | 1.157982 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.80 |
Source: Reynolds et al. (2008)