













| General Information: |
|
| Name(s) found: |
ULP2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1034 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSARKRKFNS LKPLDTLNSS RASSPRSSAS LPPKRYNTFR KDPKIVDHLN NASTKDFLPV 60
61 LSMNSESKRQ IELSDNDVDN NDEGEGVNSG CSDQDFEPLQ SSPLKRHSSL KSTSNGLLFQ 120
121 MSNNLGNGSP EPAVASTSPN GSIISTKLNL NGQFSCVDSK TLRIYRHKAP CIMTFVSDHN 180
181 HPKFSLYFQQ SVIYNSQVNL LDDVELIILD KKNSFMAIIL KDLKKVKMIL DVNNSSININ 240
241 TNILIWSTAS SASNKKIKSI KRFLLMSYSS SIKVEILDHK EQILERLKHL IHPISSSSPS 300
301 LNMERAINST KNAFDSLRLK KTKLSTNDDE SPQIHTHFLS NKPHGLQSLT KRTRIASLGK 360
361 KEHSISVPKS NISPSDFYNT NGTETLQSHA VSQLRRSNRF KDVSDPANSN SNSEFDDATT 420
421 EFETPELFKP SLCYKFNDGS SYTITNQDFK CLFNKDWVND SILDFFTKFY IESSIEKSII 480
481 KREQVHLMSS FFYTKLISNP ADYYSNVKKW VNNTDLFSKK YVVIPINISY HWFSCIITNL 540
541 DAILDFHQNK DKNDAINSDE ISINNPLVNI LTFDSLRQTH SREIDPIKEF LISYALDKYS 600
601 IQLDKTQIKM KTCPVPQQPN MSDCGVHVIL NIRKFFENPV ETIDVWKNSK IKSKHFTAKM 660
661 INKYFDKNER NSARKNLRHT LKLLQLNYIS YLKKENLYEE VMQMEEKKST NINNNENYDD 720
721 DDEEIQIIEN IDQSSKDNNA QLTSEPPCSR SSSISTTERE PTELHNSVVR QPTGEIITDN 780
781 EDPVRAASPE TASVSPPIRH NILKSSSPFI SESANETEQE EFTSPYFGRP SLKTRAKQFE 840
841 GVSSPIKNDQ ALSSTHDIMM PSPKPKRIYP SKKIPQLSSH VQSLSTDSME RQSSPNNTNI 900
901 VISDTEQDSR LGVNSESKNT SGIVNRDDSD VNLIGSSLPN VAEKNHDNTQ ESNGNNDSLG 960
961 KILQNVDKEL NEKLVDIDDV AFSSPTRGIP RTSATSKGSN AQLLSNYGDE NNQSQDSVWD1020
1021 EGRDNPILLE DEDP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
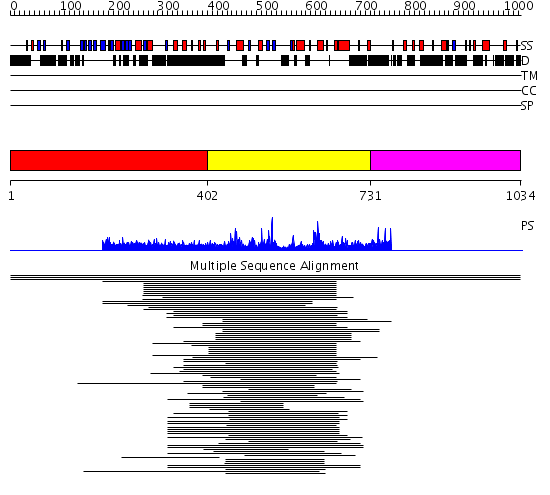
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..401] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [402..730] | 120.31133 | Ulp1 protease C-terminal domain | |
| 3 | View Details | [731..1034] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.87 |
Source: Reynolds et al. (2008)