













| General Information: |
|
| Name(s) found: |
UBP9_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 754 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MIKRWLSVNR KKSHPEKNTQ GNDEINRKAT SLKKTKGSGD PSIAKSPSAK SSTSSIPSNL 60
61 ASHERRSKFS SQTDNLAGNK HYHEHYHNMA STSDEREYDS STTYEDRAFD TESSILFTTI 120
121 TDLMPYGDGS NKVFGYENFG NTCYCNSVLQ CLYNIPEFRC NVLRYPERVA AVNRIRKSDL 180
181 KGSKIRVFTN ESFETSTNSG NSNTGYQSND NEDAHNHHHL QQSDQDNSSS STQEKQNNFE 240
241 RKRNSFMGFG KDKSNYKDSA KKDDNNEMER PQPVHTVVMA SDTLTEKLHE GCKKIIVGRP 300
301 LLKQSDSLSK ASTTDCQANS HCQCDSQGSR ITSVDDDVLV NPESCNDAVN NSNNNKENTF 360
361 PTSEQRKKAA LIRGPVLNVD HLLYPTEEAT LYNGLKDIFE SITENLSLTG IVSPTEFVKI 420
421 LKKENVLFNT MMQQDAHEFL NFLLNDFSEY IQRNNPRMRF GPQKTDNSND NFITDLFKGT 480
481 LTNRIKCLTC DNITSRDEPF LDFPIEVQGD EETDIQKMLK SYHQREMLNG VNKFYCNKCY 540
541 GLQEAERMVG LKQLPHILSL HLKRFKYSEE QKSNIKLFNK ILYPLTLDVS STFNTSVYKK 600
601 YELSGVVIHM GSGPQHGHYV CICRNEKFGW LLYDDETVES IKEETVLQFT GHPGDQTTAY 660
661 VLFYKETQAD KTENQNENID TSSQDQMQTD NNIEQLIKCD DWLRDRKLRA AANIERKKTL 720
721 GNIPEVKTAE TKTPLNDKKR NKQKRKSRIL SFIK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
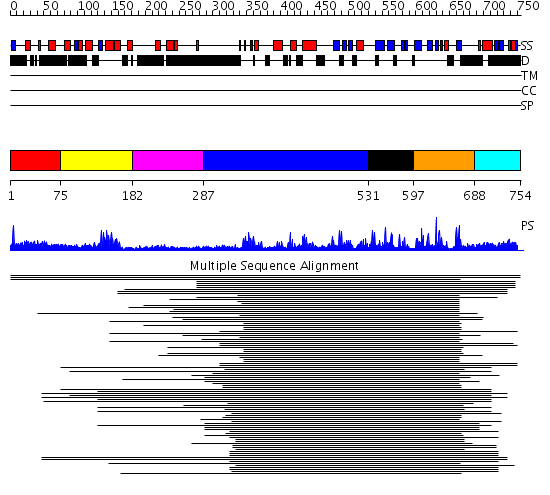
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..74] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [75..181] | 14.823909 | No description for PF00442 was found. No confident structure predictions are available. | |
| 3 | View Details | [182..286] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [287..530] | 17.244983 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [531..596] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [597..687] | 28.958607 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. | |
| 7 | View Details | [688..754] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)