













| General Information: |
|
| Name(s) found: |
UBP7_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1071 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLDDDKGTAM HPHITPFTPE YSNELLRRVQ DLYHEDIKHY YPQLKLEKLL DLLEHTEYLF 60
61 ELYLDSIHHD RPNDALTAFI IGCYYVFLII PQSLQFQTRN KSYSIYTDLK KMYENEMNMT 120
121 NVVLMVKKEI GVVLDESVKH GAGIEHRITK KRAFSVPADD LSGQVASLSL DTAAPQDHGL 180
181 KGTFTEDDAE QSSPVWTAPN LEPNDQLKLA LLPEVIPTPA FREPERKTSV PVRPSVLLED 240
241 VPSIYHEDDT SFASLNPPFR EITADRSVTH RKDSYHSVYM VDSGNLKEDN DDLFNVENDG 300
301 FIQSLDILQK QSIITAPELF SILSNRVERE KVLLIDLRIP QRSAINHIVA PNLVNVDPNL 360
361 LWDKQTNTPI YKDDILEHLL KENENFINRN KFDYIVYYTD VKTFMTINFD YAFIFFYLML 420
421 TSQKTPLTTV PTTLLGGYEK WKKTLHSYAQ EYHISIEDYL YRPYSQKARL QQEQQQQQQQ 480
481 PDSQDSFSAK ESSTKVPEPP SWKPPDLPIR LRKRPPPPPP VSMPTTPEIP PPLPPKIMVH 540
541 SQVSSISRKP PIPAKQHVKK EQLNSNEIIQ RKRQHQHQHY DQQILQPQRA YNIPTIERSP 600
601 NVYVSLSITG LRNLGNTCYI NSMIQCLFAA KTFRTLFISS KYKSYLQPIR SNGSHYSPKL 660
661 SNSLSMLFNK MYLNGGCSVV PTGFLKVINQ LRPDLKIPDD QQDTQEFLMI LLDRLHDELS 720
721 DQQHVANDYP NLLLYNADAL KVSNNEYKHW FDKNVIGNGI SPIDDIFQGQ MENSLQCKRC 780
781 GYTTFNYSTF YVLSLAIPRR SMKLSKLGRS TEKRVKLEDC INMFTSDEVL SGENAWDCPR 840
841 CGPTASVSTS VSALENEPSI VKSKKKKSRF FTLHTGTKRR HLDFFGDGIT EGHNSNNNNT 900
901 TIFERERSRS PFRMLGGSGK RSSSSTPFST GGNDSNNSSD YKNKKLTTVK TINFVTLPKI 960
961 LVIHLSRFYY DLTKKNNTVV TYPLILNIIL KNNDTMKYKL FGVVNHTGTL ISGHYTSLVN1020
1021 KDLEHNVNIG RSKWYYFDDE VVKADRKHGS DKNLKISSSD VYVLFYERVY D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
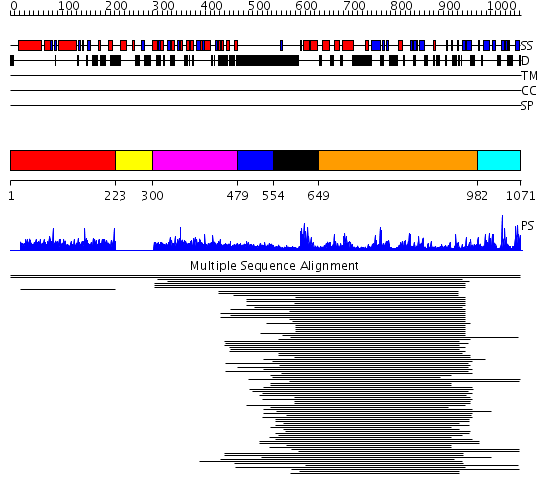
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..222] | 1.002913 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [223..299] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [300..478] | 17.29243 | Rhodanese-like domain No confident structure predictions are available. | |
| 4 | View Details | [479..553] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [554..648] | 14.236572 | No description for PF00442 was found. No confident structure predictions are available. | |
| 6 | View Details | [649..981] | 4.236992 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [982..1071] | 22.200659 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.52 |
Source: Reynolds et al. (2008)