













| General Information: |
|
| Name(s) found: |
UBP2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1272 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPNEDNELQK AIENHHNQLL NQDKENADRN GSVIEDLPLY GTSINQQSTP GDVDDGKHLL 60
61 YPDIATNLPL KTSDRLLDDI LCDTIFLNST DPKVMQKGLQ SRGILKESML SYSTFRSSIR 120
121 PNCLGSLTDQ VVFQTKSEYD SISCPKYNKI HVFQAVIFNP SLAEQQISTF DDIVKIPIYH 180
181 LKVSVKVRQE LERLKKHVGV TQFHSLDHLH EYDRVDLSTF DSSDPNLLDY GIYVSDDTNK 240
241 LILIEIFKPE FNSPEEHESF TADAIKKRYN AMCVKNESLD KSETPSQVDC FYTLFKIFKG 300
301 PLTRKSKAEP TKTIDSGNLA LNTHLNPEWL TSKYGFQASS EIDEETNEIF TEYVPPDMVD 360
361 YVNDLETRKI RESFVRKCLQ LIFWGQLSTS LLAPNSPLKN TKSVKGMSSL QTSFSTLPWF 420
421 HLLGESRARI LLNSNEQTHS PLDAEPHFIN LSVSHYYTDR DIIRNYESLS SLDPENIGLY 480
481 FDALTYIANR KGAYQLIAYC GKQDIIGQEA LENALLMFKI NPKECNISEL NEATLLSIYK 540
541 YETSNKSQVT SNHLTNLKNA LRLLAKYTKS DKLKFYVDHE PYRALSQAYD TLSIDESVDE 600
601 DIIKTAYSVK INDSPGLKLD CDRALYTIAI SKRSLDLFNF LTEECPQFSN YYGPEKLDYQ 660
661 EALKLLQVNE NASDETILKI FKQKWFDENV YEPDQFLILR AALTKISIER NSTLITNFLL 720
721 TGTIDPNSLP PENWPTGINN IGNTCYLNSL LQYYFSIAPL RRYVLEYQKT VENFNDHLSN 780
781 SGHIRRIGGR EISRGEVERS IQFIYQLRNL FYAMVHTRER CVTPSKELAY LAFAPSNVEV 840
841 EFEVEGNKVV DQTGVLSDSK KETTDDAFTT KIKDTSLIDL EMEDGLNGDV GTDANRKKNE 900
901 SNDAEVSENE DTTGLTSPTR VAKISSDQLE NALEMGRQQD VTECIGNVLF QIESGSEPIR 960
961 YDEDNEQYDL VKQLFYGTTK QSIVPLSATN KVRTKVERFL SLLINIGDHP KDIYDAFDSY1020
1021 FKDEYLTMEE YGDVIRTVAV TTFPTILQVQ IQRVYYDRER LMPFKSIEPL PFKEVIYMDR1080
1081 YADTENPLLL AKKKETEEMK QKLKVMKNRQ RELLSRDDSG LTRKDAFLES IKLLESDTIK1140
1141 KTPLKIEAAN DVIKTLRNNV QNIDNELMKL YNDINSLEEK ISHQFDDFKE YGYSLFSVFI1200
1201 HRGEASYGHY WIYIKDRNRN GIWRKYNDET ISEVQEEEVF NFNEGNTATP YFLVYVKQGQ1260
1261 EGDIEPLKRI LK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
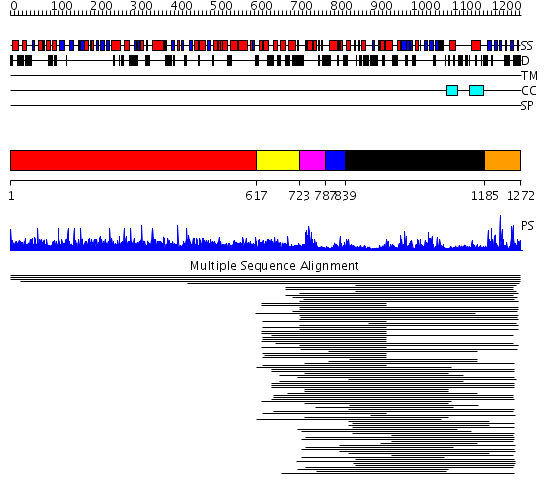
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..616] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [617..722] | 1.05699 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [723..786] | 13.69897 | No description for PF00442 was found. No confident structure predictions are available. | |
| 4 | View Details | [787..838] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [839..1184] | 14.193979 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1185..1272] | 23.886057 | Ubiquitin carboxyl-terminal hydrolase No confident structure predictions are available. | |
| 7 | View Details | [1152..1272] | 36.0 | Structure of USP14, a proteasome-associated deubiquitinating enzyme |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)