













| General Information: |
|
| Name(s) found: |
TPS2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 896 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTTTAQDNSP KKRQRIINCV TQLPYKIQLG ESNDDWKISA TTGNSALFSS LEYLQFDSTE 60
61 YEQHVVGWTG EITRTERNLF TREAKEKPQD LDDDPLYLTK EQINGLTTTL QDHMKSDKEA 120
121 KTDTTQTAPV TNNVHPVWLL RKNQSRWRNY AEKVIWPTFH YILNPSNEGE QEKNWWYDYV 180
181 KFNEAYAQKI GEVYRKGDII WIHDYYLLLL PQLLRMKFND ESIIIGYFHH APWPSNEYFR 240
241 CLPRRKQILD GLVGANRICF QNESFSRHFV SSCKRLLDAT AKKSKNSSNS DQYQVSVYGG 300
301 DVLVDSLPIG VNTTQILKDA FTKDIDSKVL SIKQAYQNKK IIIGRDRLDS VRGVVQKLRA 360
361 FETFLAMYPE WRDQVVLIQV SSPTANRNSP QTIRLEQQVN ELVNSINSEY GNLNFSPVQH 420
421 YYMRIPKDVY LSLLRVADLC LITSVRDGMN TTALEYVTVK SHMSNFLCYG NPLILSEFSG 480
481 SSNVLKDAIV VNPWDSVAVA KSINMALKLD KEEKSNLESK LWKEVPTIQD WTNKFLSSLK 540
541 EQASSNDDME RKMTPALNRP VLLENYKQAK RRLFLFDYDG TLTPIVKDPA AAIPSARLYT 600
601 ILQKLCADPH NQIWIISGRD QKFLNKWLGG KLPQLGLSAE HGCFMKDVSC QDWVNLTEKV 660
661 DMSWQVRVNE VMEEFTTRTP GSFIERKKVA LTWHYRRTVP ELGEFHAKEL KEKLLSFTDD 720
721 FDLEVMDGKA NIEVRPRFVN KGEIVKRLVW HQHGKPQDML KGISEKLPKD EMPDFVLCLG 780
781 DDFTDEDMFR QLNTIETCWK EKYPDQKNQW GNYGFYPVTV GSASKKTVAK AHLTDPQQVL 840
841 ETLGLLVGDV SLFQSAGTVD LDSRGHVKNS ESSLKSKLAS KAYVMKRSAS YTGAKV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
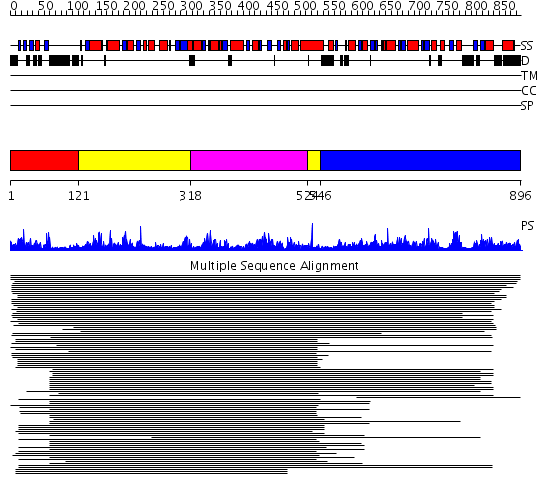
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..120] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [121..317] [524..545] |
13.39 | UDP-N-acetylglucosamine 2-epimerase | |
| 3 | View Details | [318..523] | 13.39 | UDP-N-acetylglucosamine 2-epimerase | |
| 4 | View Details | [546..896] | 25.886997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)