













| General Information: |
|
| Name(s) found: |
TOP3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 656 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKVLCVAEKN SIAKAVSQIL GGGRSTSRDS GYMYVKNYDF MFSGFPFARN GANCEVTMTS 60
61 VAGHLTGIDF SHDSHGWGKC AIQELFDAPL NEIMNNNQKK IASNIKREAR NADYLMIWTD 120
121 CDREGEYIGW EIWQEAKRGN RLIQNDQVYR AVFSHLERQH ILNAARNPSR LDMKSVHAVG 180
181 TRIEIDLRAG VTFTRLLTET LRNKLRNQAT MTKDGAKHRG GNKNDSQVVS YGTCQFPTLG 240
241 FVVDRFERIR NFVPEEFWYI QLVVENKDNG GTTTFQWDRG HLFDRLSVLT FYETCIETAG 300
301 NVAQVVDLKS KPTTKYRPLP LTTVELQKNC ARYLRLNAKQ SLDAAEKLYQ KGFISYPRTE 360
361 TDTFPHAMDL KSLVEKQAQL DQLAAGGRTA WASYAASLLQ PENTSNNNKF KFPRSGSHDD 420
421 KAHPPIHPIV SLGPEANVSP VERRVYEYVA RHFLACCSED AKGQSMTLVL DWAVERFSAS 480
481 GLVVLERNFL DVYPWARWET TKQLPRLEMN ALVDIAKAEM KAGTTAPPKP MTESELILLM 540
541 DTNGIGTDAT IAEHIDKIQV RNYVRSEKVG KETYLQPTTL GVSLVHGFEA IGLEDSFAKP 600
601 FQRREMEQDL KKICEGHASK TDVVKDIVEK YRKYWHKTNA CKNTLLQVYD RVKASM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
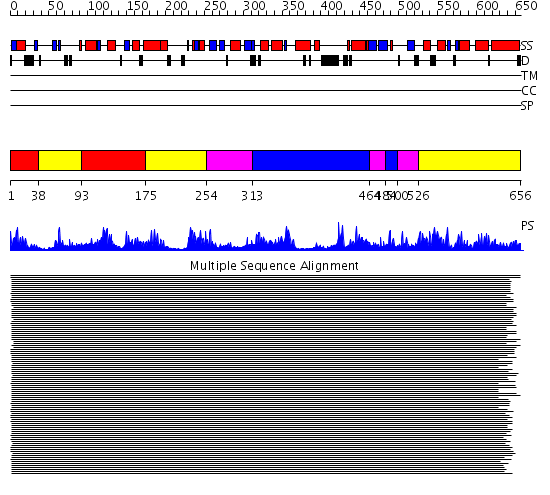
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..37] [93..174] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 2 | View Details | [38..92] [175..253] [526..656] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 3 | View Details | [254..312] [464..483] [500..525] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain | |
| 4 | View Details | [313..463] [484..499] |
408.9897 | DNA topoisomerase I, 67K N-terminal domain |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)