













| General Information: |
|
| Name(s) found: |
TIP20_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 701 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNGIDDLLNI NDRIKQVQNE RNELASKLQN LKQSLASNDT EVALSEVIAQ DIIEVGASVE 60
61 GLEQLRAKYG DLQILNKLEK VAVQQTQMQA GVDKLDSFER QLDELAEQPP DQFTLDDVKA 120
121 LHSKLTSVFA TVPQINNIDS QYAAYNKLKS KVTGKYNDVI IQRLATNWSN TFDQKLLEAQ 180
181 WDTQKFASTS VGLVKCLREN STKLYQLSLL YLPLEEETQN GDSERPLSRS NNNQEPVLWN 240
241 FKSLANNFNV RFTYHFHATS SSSKIETYFQ FLNDYLAENL YKCINIFHDD CNGLTKPVIH 300
301 EQFINYVLQP IRDKVRSTLF QNDLKTLIVL ISQILATDKN LLNSFHYHGL GLVSLISDEV 360
361 WEKWINYEVE MANRQFINIT KNPEDFPKSS QNFVKLINKI YDYLEPFYDL DFDLLVRYKL 420
421 MTCSLIFMNL TSSYLDYILT VDSLNETRTK EQELYQTMAK LQHVNFVYRK IKSLSSNFIF 480
481 IQLTDIVNST ESKKYNSLFQ NVENDYEKAM STDMQNSIVH RIQKLLKETL RNYFKISTWS 540
541 TLEMSVDENI GPSSVPSAEL VNSINVLRRL INKLDSMDIP LAISLKVKNE LLNVIVNYFT 600
601 ESILKLNKFN QNGLNQFLHD FKSLSSILSL PSHATNYKCM SLHELVKILK LKYDPNNQQF 660
661 LNPEYIKTGN FTSLKEAYSI KYLKDTKIQD ALYRIIYGNI L |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
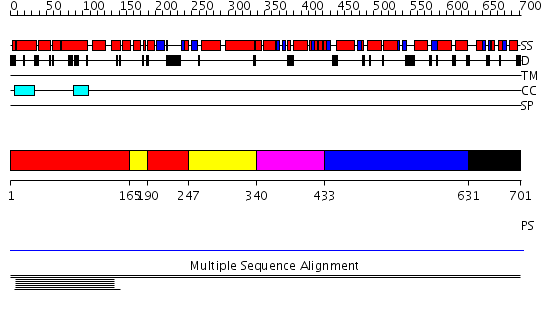
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..164] [190..246] |
7.53 | CBP80, 80KDa nuclear cap-binding protein | |
| 2 | View Details | [165..189] [247..339] |
7.53 | CBP80, 80KDa nuclear cap-binding protein | |
| 3 | View Details | [340..432] | 7.53 | CBP80, 80KDa nuclear cap-binding protein | |
| 4 | View Details | [433..630] | 7.53 | CBP80, 80KDa nuclear cap-binding protein | |
| 5 | View Details | [631..701] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||||||||||||||
| 1 |
|
|||||||||||||||||||||||||||
| 2 | No functions predicted. | |||||||||||||||||||||||||||
| 3 | No functions predicted. | |||||||||||||||||||||||||||
| 4 | No functions predicted. | |||||||||||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)