













| General Information: |
|
| Name(s) found: |
TFC3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1160 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVLTIYPDEL VQIVSDKIAS NKGKITLNQL WDISGKYFDL SDKKVKQFVL SCVILKKDIE 60
61 VYCDGAITTK NVTDIIGDAN HSYSVGITED SLWTLLTGYT KKESTIGNSA FELLLEVAKS 120
121 GEKGINTMDL AQVTGQDPRS VTGRIKKINH LLTSSQLIYK GHVVKQLKLK KFSHDGVDSN 180
181 PYINIRDHLA TIVEVVKRSK NGIRQIIDLK RELKFDKEKR LSKAFIAAIA WLDEKEYLKK 240
241 VLVVSPKNPA IKIRCVKYVK DIPDSKGSPS FEYDSNSADE DSVSDSKAAF EDEDLVEGLD 300
301 NFNATDLLQN QGLVMEEKED AVKNEVLLNR FYPLQNQTYD IADKSGLKGI STMDVVNRIT 360
361 GKEFQRAFTK SSEYYLESVD KQKENTGGYR LFRIYDFEGK KKFFRLFTAQ NFQKLTNAED 420
421 EISVPKGFDE LGKSRTDLKT LNEDNFVALN NTVRFTTDSD GQDIFFWHGE LKIPPNSKKT 480
481 PNKNKRKRQV KNSTNASVAG NISNPKRIKL EQHVSTAQEP KSAEDSPSSN GGTVVKGKVV 540
541 NFGGFSARSL RSLQRQRAIL KVMNTIGGVA YLREQFYESV SKYMGSTTTL DKKTVRGDVD 600
601 LMVESEKLGA RTEPVSGRKI IFLPTVGEDA IQRYILKEKD SKKATFTDVI HDTEIYFFDQ 660
661 TEKNRFHRGK KSVERIRKFQ NRQKNAKIKA SDDAISKKST SVNVSDGKIK RRDKKVSAGR 720
721 TTVVVENTKE DKTVYHAGTK DGVQALIRAV VVTKSIKNEI MWDKITKLFP NNSLDNLKKK 780
781 WTARRVRMGH SGWRAYVDKW KKMLVLAIKS EKISLRDVEE LDLIKLLDIW TSFDEKEIKR 840
841 PLFLYKNYEE NRKKFTLVRD DTLTHSGNDL AMSSMIQREI SSLKKTYTRK ISASTKDLSK 900
901 SQSDDYIRTV IRSILIESPS TTRNEIEALK NVGNESIDNV IMDMAKEKQI YLHGSKLECT 960
961 DTLPDILENR GNYKDFGVAF QYRCKVNELL EAGNAIVINQ EPSDISSWVL IDLISGELLN1020
1021 MDVIPMVRNV RPLTYTSRRF EIRTLTPPLI IYANSQTKLN TARKSAVKVP LGKPFSRLWV1080
1081 NGSGSIRPNI WKQVVTMVVN EIIFHPGITL SRLQSRCREV LSLHEISEIC KWLLERQVLI1140
1141 TTDFDGYWVN HNWYSIYEST |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
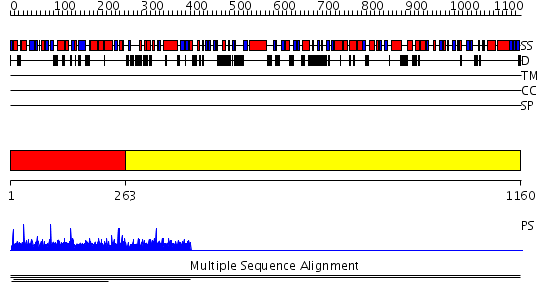
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..262] | 1.00389 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [263..1160] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)