













| General Information: |
|
| Name(s) found: |
SWE1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 819 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSLDEDEED FEMLDTENLQ FMGKKMFGKQ AGEDESDDFA IGGSTPTNKL KFYPYSNNKL 60
61 TRSTGTLNLS LSNTALSEAN SKFLGKIEEE EEEEEEGKDE ESVDSRIKRW SPFHENESVT 120
121 TPITKRSAEK TNSPISLKQW NQRWFPKNDA RTENTSSSSS YSVAKPNQSA FTSSGLVSKM 180
181 SMDTSLYPAK LRIPETPVKK SPLVEGRDHK HVHLSSSKNA SSSLSVSPLN FVEDNNLQED 240
241 LLFSDSPSSK ALPSIHVPTI DSSPLSEAKY HAHDRHNNQT NILSPTNSLV TNSSPQTLHS 300
301 NKFKKIKRAR NSVILKNREL TNSLQQFKDD LYGTDENFPP PIIISSHHST RKNPQPYQFR 360
361 GRYDNDTDEE ISTPTRRKSI IGATSQTHRE SRPLSLSSAI VTNTTSAETH SISSTDSSPL 420
421 NSKRRLISSN KLSANPDSHL FEKFTNVHSI GKGQFSTVYQ VTFAQTNKKY AIKAIKPNKY 480
481 NSLKRILLEI KILNEVTNQI TMDQEGKEYI IDYISSWKFQ NSYYIMTELC ENGNLDGFLQ 540
541 EQVIAKKKRL EDWRIWKIIV ELSLALRFIH DSCHIVHLDL KPANVMITFE GNLKLGDFGM 600
601 ATHLPLEDKS FENEGDREYI APEIISDCTY DYKADIFSLG LMIVEIAANV VLPDNGNAWH 660
661 KLRSGDLSDA GRLSSTDIHS ESLFSDITKV DTNDLFDFER DNISGNSNNA GTSTVHNNSN 720
721 INNPNMNNGN DNNNVNTAAT KNRLILHKSS KIPAWVPKFL IDGESLERIV RWMIEPNYER 780
781 RPTANQILQT EECLYVEMTR NAGAIIQEDD FGPKPKFFI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
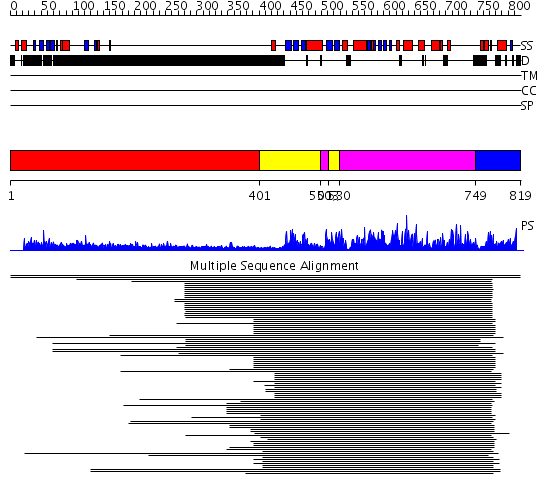
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..400] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [401..499] [513..529] |
261.218487 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [500..512] [530..748] |
261.218487 | cAMP-dependent PK, catalytic subunit | |
| 4 | View Details | [749..819] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)