













| General Information: |
|
| Name(s) found: |
SPT5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1063 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSDNSDTNVS MQDHDQQFAD PVVVPQSTDT KDENTSDKDT VDSGNVTTTE STERAESTSN 60
61 IPPLDGEEKE AKSEPQQPED NAETAATEQV SSSNGPATDD AQATLNTDSS EANEIVKKEE 120
121 GSDERKRPRE EDTKNSDGDT KDEGDNKDED DDEDDDDDDD DEDDDDEAPT KRRRQERNRF 180
181 LDIEAEVSDD EDEDEDEEDS ELVREGFITH GDDEDDEASA PGARRDDRLH RQLDQDLNKT 240
241 SEEDAQRLAK ELRERYGRSS SKQYRAAAQD GYVPQRFLLP SVDTATIWGV RCRPGKEKEL 300
301 IRKLLKKKFN LDRAMGKKKL KILSIFQRDN YTGRIYIEAP KQSVIEKFCN GVPDIYISQK 360
361 LLIPVQELPL LLKPNKSDDV ALEEGSYVRI KRGIYKGDLA MVDQISENNL EVMLKIVPRL 420
421 DYGKFDEIDP TTQQRKSRRP TFAHRAPPQL FNPTMALRLD QANLYKRDDR HFTYKNEDYI 480
481 DGYLYKSFRI QHVETKNIQP TVEELARFGS KEGAVDLTSV SQSIKKAQAA KVTFQPGDRI 540
541 EVLNGEQRGS KGIVTRTTKD IATIKLNGFT TPLEFPISTL RKIFEPGDHV TVINGEHQGD 600
601 AGLVLMVEQG QVTFMSTQTS REVTITANNL SKSIDTTATS SEYALHDIVE LSAKNVACII 660
661 QAGHDIFKVI DETGKVSTIT KGSILSKINT ARARVSSVDA NGNEIKIGDT IVEKVGSRRE 720
721 GQVLYIQTQQ IFVVSKKIVE NAGVFVVNPS NVEAVASKDN MLSNKMDLSK MNPEIISKMG 780
781 PPSSKTFQQP IQSRGGREVA LGKTVRIRSA GYKGQLGIVK DVNGDKATVE LHSKNKHITI 840
841 DKHKLTYYNR EGGEGITYDE LVNRRGRVPQ ARMGPSYVSA PRNMATGGIA AGAAATSSGL 900
901 SGGMTPGWSS FDGGKTPAVN AHGGSGGGGV SSWGGASTWG GQGNGGASAW GGAGGGASAW 960
961 GGQGTGATST WGGASAWGNK SSWGGASTWA SGGESNGAMS TWGGTGDRSA YGGASTWGGN1020
1021 NNNKSTRDGG ASAWGNQDDG NRSAWNNQGN KSNYGGNSTW GGH |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
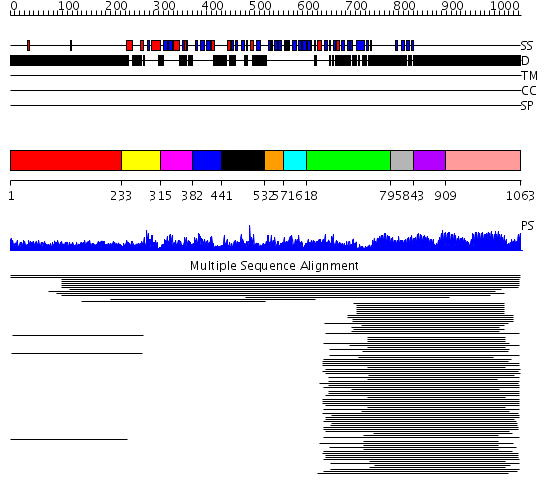
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..232] | 37.191997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [233..314] | 13.366532 | Supt5 repeat No confident structure predictions are available. | |
| 3 | View Details | [315..381] | 15.09691 | Supt5 repeat No confident structure predictions are available. | |
| 4 | View Details | [382..440] | 5.200659 | KOW motif No confident structure predictions are available. | |
| 5 | View Details | [441..531] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [532..570] | 2.744727 | KOW motif No confident structure predictions are available. | |
| 7 | View Details | [571..617] | 4.0 | KOW motif No confident structure predictions are available. | |
| 8 | View Details | [618..794] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [795..842] | 5.0 | KOW motif No confident structure predictions are available. | |
| 10 | View Details | [843..908] | N/A | No confident structure predictions are available. | |
| 11 | View Details | [909..1063] | 3.69897 | Fibrinogen |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.50 |
Source: Reynolds et al. (2008)