













| General Information: |
|
| Name(s) found: |
SPO71_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1245 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDSIVNVVED DVKYAQRVTS FSSPQNANVK VFTIPRHSFT AFRLSYVSPT ELSACSQVTL 60
61 LGGIPKQWYA DQNNQVWKLL TKISLRKVRK QSDMLRRYGY GTIYKKRVGK IPTALYLRKH 120
121 FTWSYEDNTS IHNGHRLKEA EMEMKRTRSS PVQKSEYKLS LPRRCRSSSD QNFMRQELLK 180
181 EKKSELSRNN SLPLIDTAQA VDIHPVLHEE DQENTNKRNK SLLSNLKRKD LGESKSISRK 240
241 DYSHFDRIPS PSSARSVGET DFNYNREPSE DTLRYPDSII EVTNRTSPAP NSILSRSGQF 300
301 VNSNDLSDGF STSNTINNVG LNANEKIFFN ALQSMEKENL MLWKTSQKYG TYLDERRKSA 360
361 DAFQKRERGS CVDIGKLHSS HLPFINILPP WPTELTEEER IIHDRLASKH SHHIRKHVHN 420
421 ARNKTSCKIK DSVGTFLGMT NSLTNKATVK KRTGQILKKE KMLVMVKEAI QNKVPLPNFS 480
481 ENECFDTRVS ERWKEYIVIA RSTGRFDPPI LLQFYRHRHI PEIEDISSIA TKYHRNPLDF 540
541 FLSRNCIVKF YSSLDKTISI QKPDKRLGGF IDESIEKKDE LKHYSPIKIF ILRCSSIRSS 600
601 GRWYKFLLES LDRQLFTPAI NLKIPLTEIS IKINLNEIIF QKLIDLGKQE KDRLKICFLQ 660
661 RGYKIFQHPI LRYFTVAILE KLKLAHYDYL IRKWDTENPV LGCALKRYDR LEWIPCDEDS 720
721 LVTGIFAFCQ SHLIQYRPIA NRLRETKSLE GKCLKEPTPI EGFLIRLTDK YGSARTNFGK 780
781 YSISTAYFFT CENLLFSMKA YRANPPLPID SMIDDTSTEI EKEEIWKQWK KIPEVYEQQP 840
841 YPLDTNDHIE WMNCQTTQSE YDSRDFYAFH CFHRRIDQIL KTDNVIDLTE VKDIYQGTRT 900
901 DYEADKIKYG VYKEASEIFW HRNYEIDDVS RSVINIETSN GLLLKLLATS ATVAEEWVIK 960
961 LKQMISYWKN KQREDTERLL KIRRSNAGLL MLNGEEETKI GENTLRWIVE HGRADEQTFN1020
1021 ANGISLSRPL IQKGPLYQKP HKHSVFSKYY VVLISGFIVL FHCFHRSTTG FAKEVLEYAH1080
1081 YVTIPIDDCY LYSGTTTELD LLQRDRTFDE INYGSHALPR VYGDGWRSVE DESSRCFTLW1140
1141 FGTRRALSSN RLQKKGNEKQ YTQDYGRQDN NIDPPSAPEA DLNNSNVPSN TDKIHFTKKL1200
1201 GVSGKSMVFM ARSRQERDLW VMSIYYELER LRRTASTSNS RNQTM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
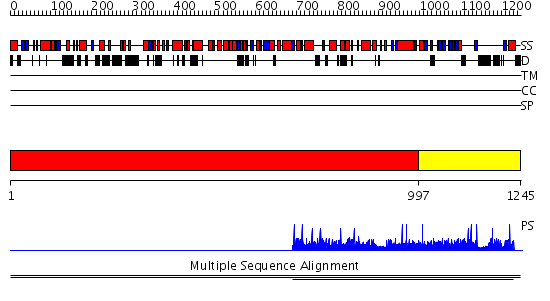
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..996] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [997..1245] | 8.60206 | PH domain No confident structure predictions are available. | |
| 3 | View Details | [439..584] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [585..694] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [695..754] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [755..844] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [845..1033] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1034..1245] | 1.07 | No description for 2dn6A was found. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.81 |
Source: Reynolds et al. (2008)