













| General Information: |
|
| Name(s) found: |
SOG2_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 791 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MVATSSKRTL DPKEEHLPAD KTSTNSSNTI ISELATQEKS SSSGTTLKLI ALNIKSISDE 60
61 DVGYIQNVER LSLRKNHLTS LPASFKRLSR LQYLDLHNNN FKEIPYILTQ CPQLEILDLS 120
121 SNEIEALPDE ISSFWQDNIR VLSLKDNNVT SIRNLKSITK LNKLSILDLE DNKIPKEELD 180
181 QVQSYTPFHT GIPKEEYWAI AISRYLKDHP NLPTPEPKIS RAAKRMGFIN TNLSNGAMNE 240
241 NNIISLAPSA NTTISASTAM VSSNQTSATS FSGTVNAESE QSGAVNGTEL YNHTKYNDYF 300
301 KRLSILPEES MSNGHQKISH AELVVSCRKL LFSFTECQQA IRKIASFCKE KAVAVNVVSL 360
361 LYSVRSHTDN LVEVLQQTEN EDESHDQALI KLCLTIITNF KQIITLLRKN FEIFFKEDDL 420
421 CFIRMFYMTL MCAYMEMYNA WSFIKEDDQV SGSASKAPKK HSFSRHETSS SSITSGGGPA 480
481 ASTTSTHCSG NIKLLPKTRS TRTPSASALL SNSNILTGDT TAVPLLSPNL NGAHTHGPIL 540
541 GHQNAISNGS SQTNMNEVKT TSDTIPRQQL LQHNKSISDS KKESQAHEPK QHPVMTSSII 600
601 NASNSNNVSN VNITPPPMNG GGAANSSANV VETNIDIQLY QTLSTVVKMV SVVYNQLTSE 660
661 ISKIAIASTM GKQILTDSLA PKIRDLTETC RQAMDLSKQL NERLNVLIPN DSNSEKYLTS 720
721 LEKLKTWEIM NSFLKVIISI LANTKIVMSD VPNLNELRPN LANLAKITKD VTVILDLSSY 780
781 KAVSVSANSP E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
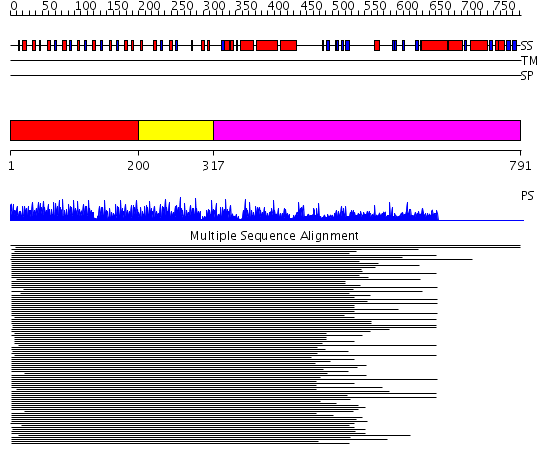
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..199] | 55.491979 | No description for 1k14A_ was found. | |
| 2 | View Details | [200..316] | 10.522879 | Rab geranylgeranyltransferase alpha-subunit, N-terminal domain; Rab geranylgeranyltransferase alpha-subunit, insert domain; Rab geranylgeranyltransferase alpha-subunit, C-terminal domain | |
| 3 | View Details | [317..791] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)