













| General Information: |
|
| Name(s) found: |
SKG3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1026 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MKRIFSGVKS PKLSAPPKVF KNDESPSTPS SPKFDQGLRS LSASASRLFS NSISTPGSPT 60
61 LDLPQEHSIN GDISPELVPI VTLLSAQAHR RYHYGIFLIL HDLKTDGTPA ARQWEECYGV 120
121 LLGTQLALWD AKELSDSKNN KNTSTMKKAA SRPSFINFTD ASVRSLDAND QVIIASENEK 180
181 TKKDLDNVLV VSTTLKNRYF LKFKNSKSFK TWNAAIRLSL FEFTALQEAY TGSFLSSRGV 240
241 KLGDIKVVMA DTKFTYEDWV SVRFGTGMPW KRCYAVISPQ SGKKKKNSKG SICFYENNKK 300
301 TKKSNIMTTV VDARALYAVY PSSPILIDTS TIIKLEGFVS FDKSEEPQET NLFIMPEKHQ 360
361 GVPGYDTIIR FLIPAMNAFY LYGRPKGLIA NRTDPDSLLF ALPTLPHIYY LQVDDVLSLT 420
421 KDKNYIHWSA ADWRNNIVQV LQKKLSKGYK GCGNKTVSVS SGMMKSPAIS SAELFEGYDS 480
481 LPERQMESPQ KSKMKSPTLA STDDINSASA SVNSHATSVK QTELFVTDNS SKINDSVSAQ 540
541 SSVTTNFKDT FTTPMTSGML NHENSERSFG SGLKLKITDS NLENMEDVEA KSANEFSTTP 600
601 EDKHIHLANA AELSALYDKY STSPFGKSEA NSSPKPQKLE VKDRSKNENR SPYERYVGTS 660
661 AESKTFEIGN VRESKSTINT SLSSPLRVED SRRSKNEDLG SLKEFEELSQ KISNMGMANI 720
721 SSEALSDTAE NSSFVTDLNL NINNSSSVNL NEEQRVPDFG EENVFDPDYM EQNQMLETES 780
781 RYTTDEFDFS DNQDAASSNY SNGQTNRTVT ETLSASDRND KIPHSSLFTN LNQLTSNGGN 840
841 YQDREDFSGD QINKPQQSQP LHVKGPQTSS FGYRNSSANS SQPQAPYPVG RPLGKIRTGP 900
901 LTVQPMQQGG NSSMYSFQSS QHRFHSSQQR QNQSLSFRNN TYGSGNNQNT FHPSPQLQQQ 960
961 PQNMRYLNNK LPINDRSPIP QTQHHVPDGR PSLHINTTNR TNPLTAQSGF SQFMPPNSTS1020
1021 TNPYSS |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
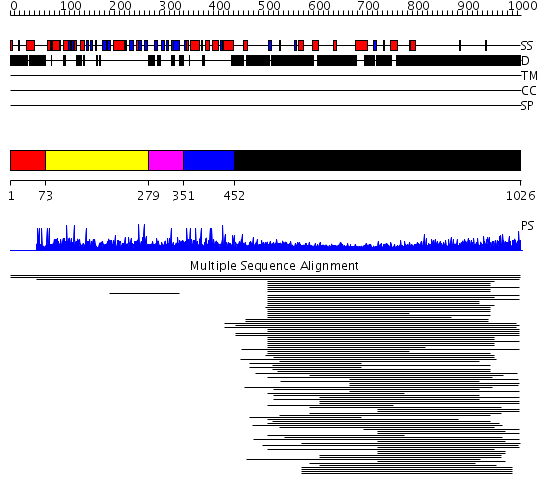
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..72] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [73..278] | 8.356547 | PH domain No confident structure predictions are available. | |
| 3 | View Details | [279..350] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [351..451] | 1.006981 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [452..1026] | 48.361998 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [741..856] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [857..1026] | 6.308997 | View MSA. No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||
| 1 | No functions predicted. | |||||||||
| 2 |
|
|||||||||
| 3 | No functions predicted. | |||||||||
| 4 | No functions predicted. | |||||||||
| 5 | No functions predicted. | |||||||||
| 6 | No functions predicted. | |||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)