













| General Information: |
|
| Name(s) found: |
SET3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 751 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSVPNSKEQS LLDDASTLLL FSKGKKRAEE ASKIGSKTDT IEHDESHERE KKGAIEMAAA 60
61 ALATASTVSL PLKKATEQSA AEAATSTAAK EETENQPQKQ PQWPVPDSYI VDPDAGIITC 120
121 ICDLNDDDGF TIQCDHCNRW QHAICYGIKD IGMAPDDYLC NSCDPREVDI NLARKIQQER 180
181 INVKTVEPSS SNNSASNKNN GRDRASSTTI SDVGDSFSTD QDNTNHRDKR RKRNPSNNSI 240
241 DSKNESASVN SSDGLTSMPK KKEHFLSAKD AYGAIYLPLK DNVFKSDLIE PFLNKHMDDN 300
301 WVIQYPHKTF KSVSIEVKPY ADIAYSRTYP GFTKLGVYLK KDCIKGDFIQ EILGELDFYK 360
361 NYLTDPRNHY RIWGTAKRRV IFHSHWPIYI DARLSGNSTR YLRRSCQPNV ELVTIKLQDT 420
421 DNRNDKSSGR KSSRIKFVLR ALRDISEDEE LYIKWQWDSK HPILKLIKGM TIDSLDDLER 480
481 YGLINSVETI LSNGECGCGN NSKDCYLLKV KRYAQSLYKS VKSRGKMNNR YKLNEILNQY 540
541 NCKKRREPPI LHRLEEKAQN TIERAPILLN NFYRQKFLNR NNGPKIPQKN TIDSTNNPDD 600
601 IAKPFKFALF AQHSSNISVP KKNETSEKPL IITKSTDYDE SHITNIEELP IPVLLPINKT 660
661 SRQTANDVEE SQSKNEHKLS RTPSLSNFNK ELSKEAQHSQ AKTKEIMTEA SVNSRRESTP 720
721 ESIMHLSDFS SSQLHSKKKL SFADYRKKLL K |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
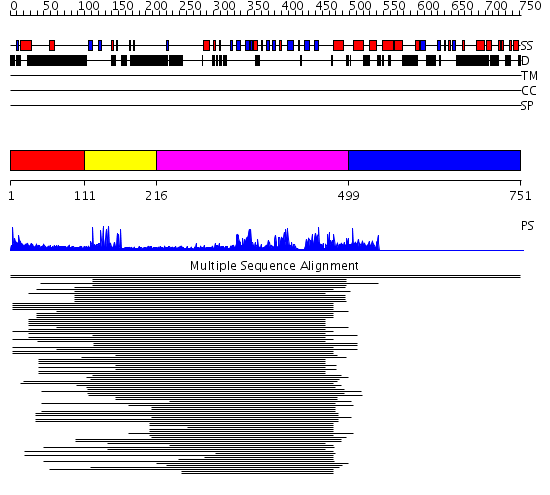
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..110] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [111..215] | 6.09691 | Nuclear corepressor KAP-1 (TIF-1beta) | |
| 3 | View Details | [216..498] | 38.169994 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [499..751] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [580..751] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)