













| General Information: |
|
| Name(s) found: |
SCY1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 804 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MMFWSSKTGI TSKYSFSSSP TFTAEPWSIY TGRPKSSSSS SPSKVSIFMF DKKQFENYLL 60
61 HYGIIKSKSG SRDKVLIQEA YEILRNQANN LAKLKHPNIL TLIEPLEEHS KNFMFVTEFV 120
121 TSSLETVFRE TDDEEQNFLQ GHVKDNIVVQ RGILQLVNAL DFVHNRASFV HLNIQPRAIF 180
181 INENSDWKIS GLGYLVKIPP GTNTSEYFLP QYDPRVPPFM HLQLNYTAPE IVFENTLTFK 240
241 NDYFSLGLLI YFLYTGKDLF RSENSTSEYK LEYNKFESKI STMSWDNIFS KVPQKLRHCI 300
301 PKLINRDIYS RYDNITLILD SEFFQDPLVK TLNFLDDLPT KNNEEKYVFL EGLVNLLPEF 360
361 PPALLQKKFL PILLELLSQF CAEKVVSDKC VGKSLDLIIK IGSTLSQLSF QEKVYPVLLS 420
421 DANFPVLLKK ATICLIDNLD TLKQKVKRSD FLENILKPLF NYVLHDSESD ITVVCQEKLL 480
481 SQIPLALEVL DFPTVKQFLL PLLSNLFTKT TSLTVKNTCV TCFQIMIEHK SIDSYTCSET 540
541 VLPLFKSMKT RDPRILSKLL KLFETVPLII TDEIVLVDQV LPLMWNYSMA STLTKSQYSG 600
601 YTKAINKMSS DIQKHHIAKL DDKVNDIGED AFHKVIEPTI MKKEDPETVA AKNIEVAAMQ 660
661 PVKKKTGSSY GETLPQSKSI LNSKPLNPKN ALATRGFPTR ILNSPPQTPS SRTGSKVMTK 720
721 GGSNDASSTK VEEEFNEFQS FSSTGSIRQT SAPSDVWMNS TPSPTPTSAS STNLPPGFSI 780
781 SLQPNKRKDG SSDIPRSNVY GSLI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
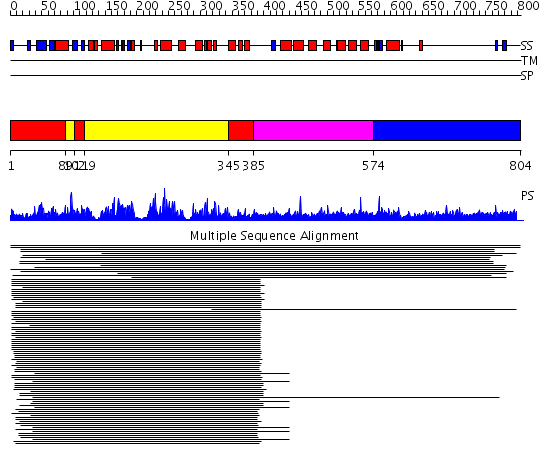
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..88] [102..118] [345..384] |
157.0 | cAMP-dependent PK, catalytic subunit | |
| 2 | View Details | [89..101] [119..344] |
157.0 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [385..573] | 2.69897 | Constant regulatory domain of protein phosphatase 2a, pr65alpha | |
| 4 | View Details | [574..804] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.91 |
Source: Reynolds et al. (2008)