













| General Information: |
|
| Name(s) found: |
SAS3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 831 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSLTANDESP KPKKNALLKN LEIDDLIHSQ FVRSDTNGHR TTRRLFNSDA SISHRIRGSV 60
61 RSDKGLNKIK KGLISQQSKL ASENSSQNIV NRDNKMGAVS FPIIEPNIEV SEELKVRIKY 120
121 DSIKFFNFER LISKSSVIAP LVNKNITSSG PLIGFQRRVN RLKQTWDLAT ENMEYPYSSD 180
181 NTPFRDNDSW QWYVPYGGTI KKMKDFSTKR TLPTWEDKIK FLTFLENSKS ATYINGNVSL 240
241 CNHNETDQEN EDRKKRKGKV PRIKNKVWFS QIEYIVLRNY EIKPWYTSPF PEHINQNKMV 300
301 FICEFCLKYM TSRYTFYRHQ LKCLTFKPPG NEIYRDGKLS VWEIDGRENV LYCQNLCLLA 360
361 KCFINSKTLY YDVEPFIFYI LTEREDTENH PYQNAAKFHF VGYFSKEKFN SNDYNLSCIL 420
421 TLPIYQRKGY GQFLMEFSYL LSRKESKFGT PEKPLSDLGL LTYRTFWKIK CAEVLLKLRD 480
481 SARRRSNNKN EDTFQQVSLN DIAKLTGMIP TDVVFGLEQL QVLYRHKTRS LSSLDDFNYI 540
541 IKIDSWNRIE NIYKTWSSKN YPRVKYDKLL WEPIILGPSF GINGMMNLEP TALADEALTN 600
601 ETMAPVISNN THIENYNNSR AHNKRRRRRR RSSEHKTSKL HVNNIIEPEV PATDFFEDTV 660
661 SSLTEYMCDY KNTNNDRLIY QAEKRVLESI HDRKGIPRSK FSTETHWELC FTIKNSETPL 720
721 GNHAARRNDT GISSLEQDEV ENDVDTELYV GENAKEDEDE DEDFTLDDDI EDEQISEEND 780
781 EEEDTYEEDS DDDEDGKRKG QEQDENDIES HIRKERVRKR RKITLIEDDE E |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
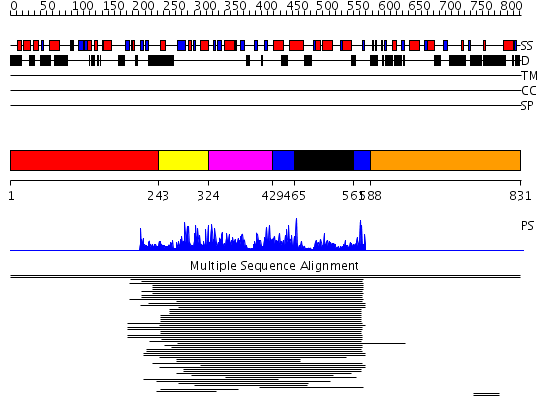
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..242] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [243..323] | 730.0 | Histone acetyltransferase ESA1 | |
| 3 | View Details | [324..428] | 730.0 | Histone acetyltransferase ESA1 | |
| 4 | View Details | [429..464] [561..587] |
730.0 | Histone acetyltransferase ESA1 | |
| 5 | View Details | [465..560] | 730.0 | Histone acetyltransferase ESA1 | |
| 6 | View Details | [588..831] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)