













| General Information: |
|
| Name(s) found: |
SAC3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1301 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNTSFGSVVP STNFNFFKGH GNNDNTSANS TVNNSNFFLN SNETKPSKNV FMVHSTSQKK 60
61 SQQPLQNLSH SPSYTENKPD KKKKYMINDA KTIQLVGPLI SSPDNLGFQK RSHKARELPR 120
121 FLINQEPQLE KRAFVQDPWD KANQEKMISL EESIDDLNEL YETLKKMRNT ERSIMEEKGL 180
181 VDKADSAKDL YDAIVFQGTC LDMCPTFERS RRNVEYTVYS YEKNQPNDKK ASRTKALKVF 240
241 ARPAAAAAPP LPSDVRPPHI LVKTLDYIVD NLLTTLPESE GFLWDRMRSI RQDFTYQNYS 300
301 GPEAVDCNER IVRIHLLILH IMVKSNVEFS LQQELEQLHK SLITLSEIYD DVRSSGGTCP 360
361 NEAEFRAYAL LSKIRDPQYD ENIQRLPKHI FQDKLVQMAL CFRRVISNSA YTERGFVKTE 420
421 NCLNFYARFF QLMQSPSLPL LMGFFLQMHL TDIRFYALRA LSHTLNKKHK PIPFIYLENM 480
481 LLFNNRQEII EFCNYYSIEI INGDAADLKT LQHYSHKLSE TQPLKKTYLT CLERRLQKTT 540
541 YKGLINGGED NLASSVYVKD PKKDRIPSIA DQSFLMENFQ NNYNEKLNQN SSVKPQINTS 600
601 PKRVATRPNH FPFSQESKQL PQISQSHTLS TNPLLTPQVH GDLSEQKQQQ IKTVTDGGSP 660
661 FVFDQSAQNS TVEASKAHMI STTSNGAYDE KLSSEQEEMR KKEEQRIEEE KTQLKKKQEN 720
721 ADKQVITEQI ANDLVKEVVN SSVISIVKRE FSEANYRKDF IDTMTRELYD AFLHERLYLI 780
781 YMDSRAELKR NSTLKKKFFE KWQASYSQAK KNRILEEKKR EEIKLVSHQL GVPGFKKSTC 840
841 LFRTPYKGNV NSSFMLSSSD KNLIFSPVND EFNKFATHLT KISKLWRPLE MQSIYYDNLT 900
901 KKFPSNSLTP ANLFIYAKDW TSLSNRWILS KFNLQTAQDS KKFSNNIISS RIICIDDEYE 960
961 PSDFSDLQLL IFNTGVTNPD IFDLEMKLKD DGEELIKLIT GISLNTNICF SLLIIYWESA1020
1021 ENTLSESTIK HLLKLNRISK NYSSVIERID LMNLTEESPH KCLEDKLSEI SHSYVYKLTE1080
1081 RGKYDKTLRQ KRSLAGIHSR STQLQTTKDI DQKMKKMLEK EKNKYQQQIG ERNTYAHLES1140
1141 HIDASPRSKK RKLPILLSTS HSSQFKTPLA SRLNTSGSST SPPLPSHLAM KFRKNSRVTS1200
1201 LHTVLPVSTP SHSNNIPAAS FSGNNTTDIQ SQQLIENQKS TSVYLNNVSE RILGNQEICQ1260
1261 TPINPVTPVL DGADQGKEDI PDSILELKIL IDSVKKKVNN D |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
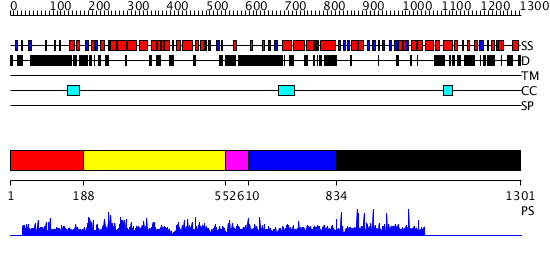
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..187] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [188..551] | 140.017729 | SAC3/GANP/Nin1/mts3/eIF-3 p25 family No confident structure predictions are available. | |
| 3 | View Details | [552..609] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [610..833] | 3.221849 | Heavy meromyosin subfragment | |
| 5 | View Details | [834..1301] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)