













| General Information: |
|
| Name(s) found: |
RIR1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 888 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MYVYKRDGRK EPVQFDKITA RISRLCYGLD PKHIDAVKVT QRIISGVYEG VTTIELDNLA 60
61 AETCAYMTTV HPDYATLAAR IAISNLHKQT TKQFSKVVED LYRYVNAATG KPAPMISDDV 120
121 YNIVMENKDK LNSAIVYDRD FQYSYFGFKT LERSYLLRIN GQVAERPQHL IMRVALGIHG 180
181 RDIEAALETY NLMSLKYFTH ASPTLFNAGT PKPQMSSCFL VAMKEDSIEG IYDTLKECAL 240
241 ISKTAGGIGL HIHNIRSTGS YIAGTNGTSN GLIPMIRVFN NTARYVDQGG NKRPGAFALY 300
301 LEPWHADIFD FIDIRKNHGK EEIRARDLFP ALWIPDLFMK RVEENGTWTL FSPTSAPGLS 360
361 DCYGDEFEAL YTRYEKEGRG KTIKAQKLWY SILEAQTETG TPFVVYKDAC NRKSNQKNLG 420
421 VIKSSNLCCE IVEYSAPDET AVCNLASVAL PAFIETSEDG KTSTYNFKKL HEIAKVVTRN 480
481 LNRVIDRNYY PVEEARKSNM RHRPIALGVQ GLADTFMLLR LPFDSEEARL LNIQIFETIY 540
541 HASMEASCEL AQKDGPYETF QGSPASQGIL QFDMWDQKPY GMWDWDTLRK DIMKHGVRNS 600
601 LTMAPMPTAS TSQILGYNEC FEPVTSNMYS RRVLSGEFQV VNPYLLRDLV DLGIWDEGMK 660
661 QYLITQNGSI QGLPNVPQEL KDLYKTVWEI SQKTIINMAA DRSVYIDQSH SLNLFLRAPT 720
721 MGKLTSMHFY GWKKGLKTGM YYLRTQAASA AIQFTIDQKI ADQATENVAD ISNLKRPSYM 780
781 PSSASYAASD FVPAAVTANA TIPSLDSSSE ASREASPAPT GSHSLTKGMA ELNVQESKVE 840
841 VPEVPAPTKN EEKAAPIVDD EETEFDIYNS KVIACAIDNP EACEMCSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
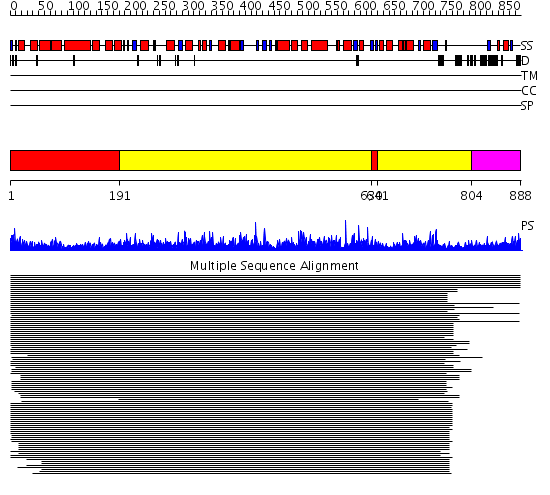
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..190] [630..640] |
1616.9897 | R1 subunit of ribonucleotide reductase, N-terminal domain; R1 subunit of ribonucleotide reductase, C-terminal domain | |
| 2 | View Details | [191..629] [641..803] |
1616.9897 | R1 subunit of ribonucleotide reductase, N-terminal domain; R1 subunit of ribonucleotide reductase, C-terminal domain | |
| 3 | View Details | [804..888] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)