













| General Information: |
|
| Name(s) found: |
RAP1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 827 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSSPDDFETA PAEYVDALDP SMVVVDSGSA AVTAPSDSAA EVKANQNEEN TGATAAETSE 60
61 KVDQTEVEKK DDDDTTEVGV TTTTPSIADT AATANIASTS GASVTEPTTD DTAADEKKEQ 120
121 VSGPPLSNMK FYLNRDADAH DSLNDIDQLA RLIRANGGEV LDSKPRESKE NVFIVSPYNH 180
181 TNLPTVTPTY IKACCQSNSL LNMENYLVPY DNFREVVDSR LQEESHSNGV DNSNSNSDNK 240
241 DSIRPKTEII STNTNGATED STSEKVMVDA EQQARLQEQA QLLRQHVSST ASITSGGHND 300
301 LVQIEQPQKD TSNNNNSNVN DEDNDLLTQD NNPQTADEGN ASFQAQRSMI SRGALPSHNK 360
361 ASFTDEEDEF ILDVVRKNPT RRTTHTLYDE ISHYVPNHTG NSIRHRFRVY LSKRLEYVYE 420
421 VDKFGKLVRD DDGNLIKTKV LPPSIKRKFS ADEDYTLAIA VKKQFYRDLF QIDPDTGRSL 480
481 ITDEDTPTAI ARRNMTMDPN HVPGSEPNFA AYRTQSRRGP IAREFFKHFA EEHAAHTENA 540
541 WRDRFRKFLL AYGIDDYISY YEAEKAQNRE PEPMKNLTNR PKRPGVPTPG NYNSAAKRAR 600
601 NYSSQRNVQP TANAASANAA AAAAAAASNS YAIPENELLD EDTMNFISSL KNDLSNISNS 660
661 LPFEYPHEIA EAIRSDFSNE DIYDNIDPDT ISFPPKIATT DLFLPLFFHF GSTRQFMDKL 720
721 HEVISGDYEP SQAEKLVQDL CDETGIRKNF STSILTCLSG DLMVFPRYFL NMFKDNVNPP 780
781 PNVPGIWTHD DDESLKSNDQ EQIRKLVKKH GTGRMEMRKR FFEKDLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
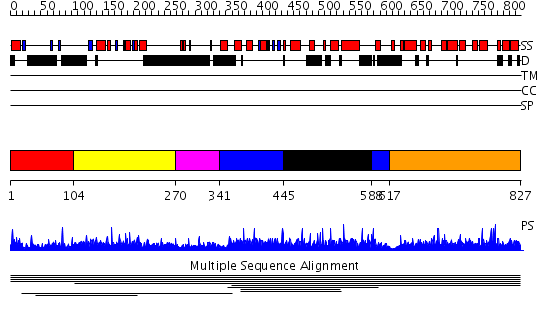
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..103] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [104..269] | 7.52 | DNA ligase III alpha | |
| 3 | View Details | [270..340] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [341..444] [588..616] |
1518.0 | DNA-binding domain of rap1 | |
| 5 | View Details | [445..587] | 1518.0 | DNA-binding domain of rap1 | |
| 6 | View Details | [617..827] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)