













| General Information: |
|
| Name(s) found: |
PPX1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 397 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSPLRKTVPE FLAHLKSLPI SKIASNDVLT ICVGNESADM DSIASAITYS YCQYIYNEGT 60
61 YSEEKKKGSF IVPIIDIPRE DLSLRRDVMY VLEKLKIKEE ELFFIEDLKS LKQNVSQGTE 120
121 LNSYLVDNND TPKNLKNYID NVVGIIDHHF DLQKHLDAEP RIVKVSGSCS SLVFNYWYEK 180
181 LQGDREVVMN IAPLLMGAIL IDTSNMRRKV EESDKLAIER CQAVLSGAVN EVSAQGLEDS 240
241 SEFYKEIKSR KNDIKGFSVS DILKKDYKQF NFQGKGHKGL EIGLSSIVKR MSWLFNEHGG 300
301 EADFVNQCRR FQAERGLDVL VLLTSWRKAG DSHRELVILG DSNVVRELIE RVSDKLQLQL 360
361 FGGNLDGGVA MFKQLNVEAT RKQVVPYLEE AYSNLEE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
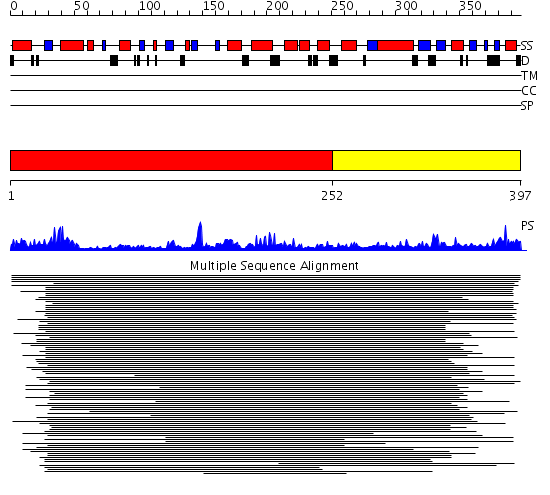
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..251] | 56.522879 | Manganese-dependent inorganic pyrophosphatase (family II) | |
| 2 | View Details | [252..397] | 56.522879 | Manganese-dependent inorganic pyrophosphatase (family II) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||||||||||||||
| 2 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)