













| General Information: |
|
| Name(s) found: |
PMD1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1753 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVLQPPSSV CYPLNLPIVP NPNLDEATRK KLTLECRTGA AVELARSGVF VHGGLTLPLN 60
61 LTIINSLQLQ KELILYFGKQ KDRNADFKTL ADWISPEIFF LDLISRTWQR INTTIDTTSE 120
121 NELNNGLSFK ERLFHSMCFT ESNIYIFGGL MVSPHNGYEL IATNELWKLD LKTKCWSLIS 180
181 ENPQITRRFN HSMHVLNENN ENQDTKLIIV GGLDNMDIPV KKIDIFNLRT SLWESESKSD 240
241 ENPASKGSSK ILVNIDGMPI SLSHDSNFSV LIENNQAEIP TLALYYPQRE ANTSRRGTDD 300
301 GSFSTYAHDL DDKSKLPKHH HHHHGDLKYF ESDDADENAV KTLMSPIVIL PLLGNSQGAR 360
361 MTSNPTQNNK ENSILQVPFH LQYPSGNYFN YNIVVIGFYP DPQPSNLHCF IYNIASGKWI 420
421 RVNIACTECS ISMHRFWKLL IWKSHHQALL LGTRTDDFCS PSVQKFDHIL SFSLPMLNGY 480
481 NKLVNTKHTR TNNGIANSHN LNVNLSLYDH LPYSNSSTIE HTNPYTVTQG YSLDDSGIPR 540
541 LTSTATSQFE NYSRYITVPL EMESTSSIFP PYAMVLGKDA LEIFGKTLSD FEFITADGDS 600
601 IGVPVYLLRK RWGRYFDSLL SNGYANTSFN YEFNGDTSNI ISFSPHTASK TTKFGNSSQS 660
661 SNGSLEKYFS KNGNSKSNSN TSLKKPHSVD FTSSTSSPKQ RAISHNKLSP SEPILCADEE 720
721 DSRSNTLKQH ATGDTGLKET GTSNKRPIST TCSSTGMVFR VPFQDMKNSK LGLSEQSGRS 780
781 TRASSVSPPP VYKKSTNDGN DSNCTLSNTP LVYRRASTVG TTTNSSVDDG FSSIRRASHP 840
841 LQSYIIAKSS PSSISKASPA EKAFSRRKSS ALRFIASPNQ SRQTSFASTA STASVVSSTS 900
901 GRRRNSNQIS HLGSSASLPN SPILPVLNIP LPPQEKIPLE PLPPVPKAPS RRSSSLAEYV 960
961 QFGRDSPVAS RRSSHSTRKS SSSDARRISN SSLLRNTLDS QLLSNSYGSD IPYEASIQEY1020
1021 GMNNGRDEEE DGDNQDYGCI SPSNIRPIFS TINAININGN FKEGEFFSSK SYINNEKSRR1080
1081 LSYISNPESV ESTNSNNNAI IELEPLLTPR SLYMPWSTAS VRAFAEFFYT AQINGKWLLA1140
1141 PVTLDLLIMA KIYEIPILYE LITEVLYKII SKKEEGLSVT CEALLNLFQQ KVSRYCNENE1200
1201 GKIRKQLDSS ESYQDTLEIK RSLANIDNGY VDSYLLRNTS MAQSIHYTDD SNGETEIDMH1260
1261 HTGISSIGSL ANRAVPTVFA GGPRDSHNSI GSIAFPSNSG VQNIRRSVSL FSPATKKKSS1320
1321 LSRETDPLDT SDQFTDDVPD SGPVSRQQNF PRRSSSFTET VPTEPTRYNY QNLDSSKSNR1380
1381 ASDDKEEQNE QATLQDISNF DKYKVETLQK RNSNDGKDLD RTNDPLKNRG TEIPQNSSNL1440
1441 ETDPFIRDSF DSDSGSSFRS DSDDLDSQLG ILPFTKMNKK LQEQTSQEFD DSIDPLYKIG1500
1501 SSTPGSSRLH GSFSKYIRPN SQREDGSEYV NISSLENMVS PNALPPVDYV MKSIYRTSVL1560
1561 VNDSNLMTRT KEAIELSKVL KKLKKKVLQD ISQMDDEMRE TGKPIFARGS SSPTLSRQHS1620
1621 DVATPLKQQE NTRPALKFAS SSPISEGFRK SSIKFSQAPS TQISPRTSVT DFTASQQRRQ1680
1681 HMNKRFSTQT THSTSALFMN PAFMPSAVNT GRKESEGHCE DRSATANRTN RKEDATTNDN1740
1741 DNIAPFPFFG KRR |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
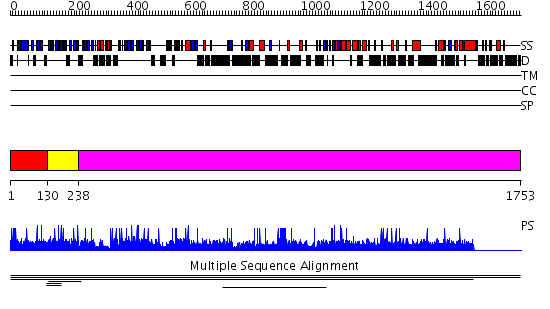
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..129] | N/A | Confident ab initio structure predictions are available. | |
| 2 | View Details | [130..237] | 5.356547 | Kelch motif No confident structure predictions are available. | |
| 3 | View Details | [238..1753] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [727..945] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [946..1095] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [1096..1299] | N/A | No confident structure predictions are available. | |
| 7 | View Details | [1300..1433] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [1434..1666] | N/A | No confident structure predictions are available. | |
| 9 | View Details | [1667..1753] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.86 |
Source: Reynolds et al. (2008)