













| General Information: |
|
| Name(s) found: |
PEX5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDVGSCSVGN NPLAQLHKHT QQNKSLQFNQ KNNGRLNESP LQGTNKPGIS EAFISNVNAI 60
61 SQENMANMQR FINGEPLIDD KRRMEIGPSS GRLPPFSNVH SLQTSANPTQ IKGVNDISHW 120
121 SQEFQGSNSI QNRNADTGNS EKAWQRGSTT ASSRFQYPNT MMNNYAYASM NSLSGSRLQS 180
181 PAFMNQQQSG RSKEGVNEQE QQPWTDQFEK LEKEVSENLD INDEIEKEEN VSEVEQNKPE 240
241 TVEKEEGVYG DQYQSDFQEV WDSIHKDAEE VLPSELVNDD LNLGEDYLKY LGGRVNGNIE 300
301 YAFQSNNEYF NNPNAYKIGC LLMENGAKLS EAALAFEAAV KEKPDHVDAW LRLGLVQTQN 360
361 EKELNGISAL EECLKLDPKN LEAMKTLAIS YINEGYDMSA FTMLDKWAET KYPEIWSRIK 420
421 QQDDKFQKEK GFTHIDMNAH ITKQFLQLAN NLSTIDPEIQ LCLGLLFYTK DDFDKTIDCF 480
481 ESALRVNPND ELMWNRLGAS LANSNRSEEA IQAYHRALQL KPSFVRARYN LAVSSMNIGC 540
541 FKEAAGYLLS VLSMHEVNTN NKKGDVGSLL NTYNDTVIET LKRVFIAMNR DDLLQEVKPG 600
601 MDLKRFKGEF SF |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
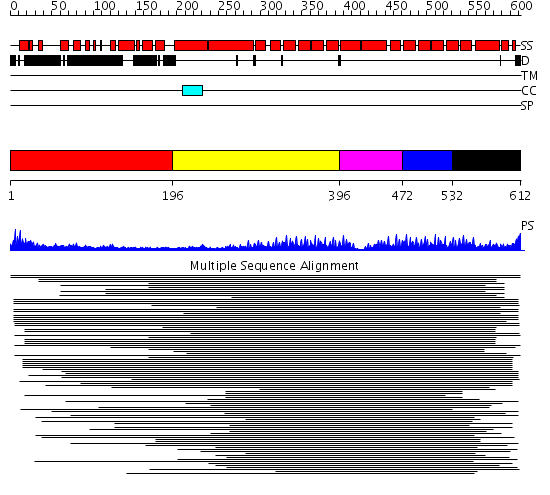
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..195] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [196..395] | 525.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [396..471] | 525.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [472..531] | 525.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 5 | View Details | [532..612] | 525.221849 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||
| 1 | No functions predicted. | ||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||
| 3 |
|
||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||
| 5 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)