













| General Information: |
|
| Name(s) found: |
PAH1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 862 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQYVGRALGS VSKTWSSINP ATLSGAIDVI VVEHPDGRLS CSPFHVRFGK FQILKPSQKK 60
61 VQVFINEKLS NMPMKLSDSG EAYFVFEMGD QVTDVPDELL VSPVMSATSS PPQSPETSIL 120
121 EGGTEGEGEG ENENKKKEKK VLEEPDFLDI NDTGDSGSKN SETTGSLSPT ESSTTTPPDS 180
181 VEERKLVEQR TKNFQQKLNK KLTEIHIPSK LDNNGDLLLD TEGYKPNKNM MHDTDIQLKQ 240
241 LLKDEFGNDS DISSFIKEDK NGNIKIVNPY EHLTDLSPPG TPPTMATSGS VLGLDAMESG 300
301 STLNSLSSSP SGSDTEDETS FSKEQSSKSE KTSKKGTAGS GETEKRYIRT IRLTNDQLKC 360
361 LNLTYGENDL KFSVDHGKAI VTSKLFVWRW DVPIVISDID GTITKSDALG HVLAMIGKDW 420
421 THLGVAKLFS EISRNGYNIL YLTARSAGQA DSTRSYLRSI EQNGSKLPNG PVILSPDRTM 480
481 AALRREVILK KPEVFKIACL NDIRSLYFED SDNEVDTEEK STPFFAGFGN RITDALSYRT 540
541 VGIPSSRIFT INTEGEVHME LLELAGYRSS YIHINELVDH FFPPVSLDSV DLRTNTSMVP 600
601 GSPPNRTLDN FDSEITSGRK TLFRGNQEEK FTDVNFWRDP LVDIDNLSDI SNDDSDNIDE 660
661 DTDVSQQSNI SRNRANSVKT AKVTKAPQRN VSGSTNNNEV LAASSDVENA SDLVSSHSSS 720
721 GSTPNKSTMS KGDIGKQIYL ELGSPLASPK LRYLDDMDDE DSNYNRTKSR RASSAAATSI 780
781 DKEFKKLSVS KAGAPTRIVS KINVSNDVHS LGNSDTESRR EQSVNETGRN QLPHNSMDDK 840
841 DLDSRVSDEF DDDEFDEDEF ED |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
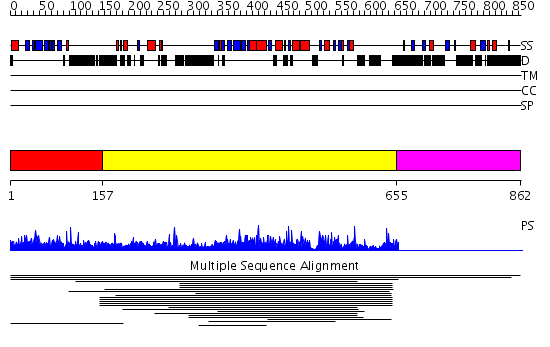
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..156] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [157..654] | 4.01789 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [655..862] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [657..862] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)