













| General Information: |
|
| Name(s) found: |
OPLA_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1286 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MQKGNIRIAI DKGGTFTDCV GNIGTGKQEH DTVIKLLSVD PKNYPDAPLE GIRRLLEVLE 60
61 HKTIPRGIPL DISNVRSLRM GTTLATNCAL ERNGERCAFI TTKGFKDSLL IGDQTRPDIF 120
121 NLNIKKVVPL YDTVVEIDER VTLEDFSEDP YFTKSSPNEQ EGILEGNSGE MVRVIKKPDE 180
181 SSVRSILKVL YASGIKSIAI AFLHSYTFPD HERIVGNIAR EIGFSHVSLS SEVSPMIKFL 240
241 PRAHSSVADA YLTPVIKKYL NSISAGLSHA EDTHIQFMQS DGGLVDGGKF SGLKSILSGP 300
301 AGGVIGYSST CYDKNNNIPL IGFDMGGTST DVSRYGDGRL EHVFETVTAG IIIQSPQLDI 360
361 HTVAAGGSSI LSWKNGLFRV GPDSAAADPG PAAYRKGGPL TITDANLFLG RLVPEFFPKI 420
421 FGPNEDESLD LETTTLKFRE LTDVINKDLN SNLTMEEVAY GFIKVANECM ARPVRAITEA 480
481 KGHVVSQHRL VSFGGAGGQH AIAVADSLGI DTVLIHRYSS ILSAYGIFLA DVIEENQEPC 540
541 SFILGEPETI LKVKKRFLEL SKNSIKNLLS QSFSREDIVL ERYLNLRYEG TETSLMILQK 600
601 YDDQWNFREW FSEAHKKEFG FSFDDKRIII DDIRIRAIGK SGVRKEKTVD EQLIEISHFK 660
661 KADVSKDASF TQKAYFDNKW VDTAVFKIDD LPAGTIIEGP AILADGTQTN IILPNSQATI 720
721 LNSHIFIKIN QKAAKTLSKS GYELDIDPIL LSIFSHRFMD IALQMGTQLR KTSVSTNVKE 780
781 RLDFSCALFD SKGNLVANAP HVPVHLGSMS TCISAQAKLW EGKLKPGDVL ITNHPDIGGT 840
841 HLPDITVITP SFSSTGELIF YVASRAHHAD IGGILPGSVP PNSKELYEEG TAIYSELVVK 900
901 EGIFQEELIY KLFVEDPGKY PGCSGSRRFS DNISDLKAQV AANTKGIQLI GSLTKEYDLA 960
961 TILKYMAAIQ TNASESIKKM LAKMVEHFGT TKFSGEDRLD DGSLIKLQVI IRPEKEEYIF1020
1021 NFDGTSPQVY GNLNAPEAIT NSAILYCLRC LVGEDIPLNQ GCLKPLTIKI PAGSLLSPRS1080
1081 GAAVVGGNVL TSQRVTDVIL KTFNVMADSQ GDCNNFTFGT GGNSGNKTDK QIKGFGYYET1140
1141 ICGGSGAGAD SWRGSGWNGS DAVHTNMTNT RMTDTEVFER RYPVLLKEFS IRRGSGGKGK1200
1201 YTGGNGVVRD VQFRKAVTAS ILSERRVIGP HGIKGGQDGS RGENLWVRHS TGALINVGGK1260
1261 NTIYAQPGDR FIIKTPGGGG FGQYKD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
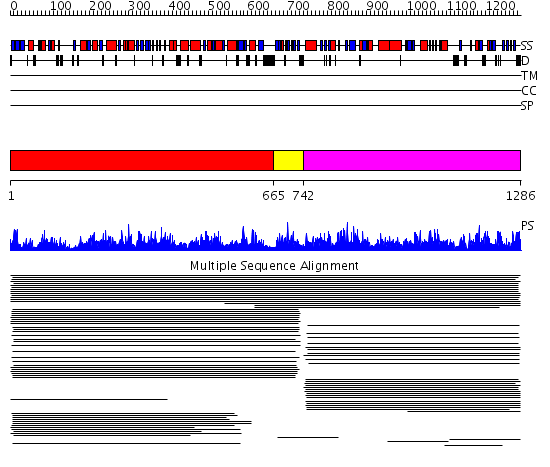
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..664] | 1000.0 | Hydantoinase/oxoprolinase No confident structure predictions are available. | |
| 2 | View Details | [665..741] | N/A | Confident ab initio structure predictions are available. | |
| 3 | View Details | [742..1286] | 1000.0 | Hydantoinase B/oxoprolinase No confident structure predictions are available. | |
| 4 | View Details | [878..957] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [958..1119] | 1.177994 | View MSA. No confident structure predictions are available. | |
| 6 | View Details | [1120..1181] | 1.176993 | View MSA. No confident structure predictions are available. | |
| 7 | View Details | [1182..1286] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||||||||||||||||||||
| 1 |
|
||||||||||||||||||||||||||||||
| 2 | No functions predicted. | ||||||||||||||||||||||||||||||
| 3 | No functions predicted. | ||||||||||||||||||||||||||||||
| 4 | No functions predicted. | ||||||||||||||||||||||||||||||
| 5 | No functions predicted. | ||||||||||||||||||||||||||||||
| 6 | No functions predicted. | ||||||||||||||||||||||||||||||
| 7 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)