













| General Information: |
|
| Name(s) found: |
OAF3_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 863 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MGYDSQVRTK KRHRITVVCT NCKKRKSKCD RTKPCGTCVR LGDVDSCVYL TDSSGQPESS 60
61 PSLNDADPLR KQSTPAERIS PGFIKKRRSS QTRQDEDHWQ RVRELENQSS LYYLPIHEET 120
121 PFFIDLIPNG FYLETKRSAD NLFGLFTDRA IENRDPYLQA MVTFRSIAIK KMMDKLGSNG 180
181 NNVKNGSLPK SFEALSTFDA DDERHISDDV VGKGNNFRMH QTIHKSLFNK FAQYRENNAK 240
241 KFSSETILAK DYLPPLKILE SEVLALFEEK IYNMIPIFDM KVLRHEITIF YQNIVEKGNP 300
301 ISIKHYDHMV FCIILLIIKI CRLSVQFSKL TPYIYPVLQE IDTSKFLALV KHYLFETKVL 360
361 RKCNLLQLQC LILLRFLHWC APEDGDGPET QYCQILMGTI ISSCKEMGIN WYCFSHPEKY 420
421 SFKINRHTRP SYDIMKPSDY ISVFRKIWSY VLFWDRKMCF ISGEECQIGK TLQCHFKEEA 480
481 DTPTWYIRML TLDNLMKKIN DTLNDDPGKV DLNLLHRLIN DLKRNFHILK SLSKNEKETM 540
541 RHFDFEMEWI IDLFSLSLLH GEMIFYEYDC NITKFYKSFQ DLWDMVIHIS EKCYNYFFNS 600
601 DALEVDSLTK FYTNRIVEIV ANKVLVIVPA FILRGDRFKT IQYADKKKMI EFLYGVSSVY 660
661 FNEFGFEYYR CFRKMFTAKI AYKILNRSCE KDAWRIILKF LLNELKLEDN GDSYIDYNDM 720
721 RLNDICPIIL EFQETVQKYD GYRPDILSIW NNEFYPIGKY NDDMTGFKFQ MRIKEMQEFL 780
781 DMEKYSDRFN IFSSFYDHAS SQLAKHTEVD TNISITNEQV AEIPQKELLQ QPLAPALPVN 840
841 DLIVSEFDVI EDIFDPVDFV SFF |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
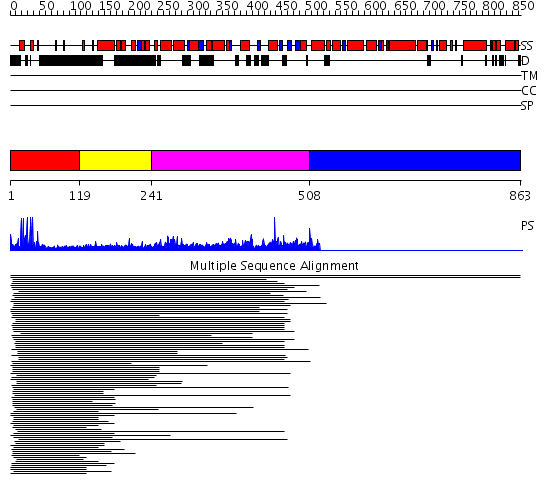
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] | 51.045757 | Hap1 (Cyp1); HAP1 | |
| 2 | View Details | [119..240] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [241..507] | 2.052995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [508..863] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)