













| General Information: |
|
| Name(s) found: |
NTO1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 748 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNRGSLDDGP KLREEKHFQD FYPDLNADTL LPFIVPLVET KDNSTDTDSD DISNRNNREI 60
61 GSVKSVQTKE LIFKGRVTTE PLVLKKNEVE FQKCKITTNE LKGKKNPYCV RFNESFISRY 120
121 YHINKVRNRK SYKQQQKEFD GVEAPYFTKF SSKEAPNITI STSTKSAIQK FASISPNLVN 180
181 FKPQYDMDEQ DELYLHYLNK RYFKDQMSHE IFEILMTTLE TEWFHIEKHI PSTNSLIARH 240
241 NILRDCKNYE LYGSDDGTGL SMDQACAVCL GTDSDNLNTI VFCDGCDIAV HQECYGIIFI 300
301 PEGKWLCRRC MISKNNFATC LMCPSHTGAF KQTDTGSWVH NICALWLPEL YFSNLHYMEP 360
361 IEGVQNVSVS RWKLNCYICK KKMGACIQCF QRNCFTAYHV TCARRAGLYM SKGKCTIQEL 420
421 ASNQFSQKYS VESFCHKHAP RGWQTSIEGI NKARKYFSLL STLQTETPQH NEANDRTNSK 480
481 FNKTIWKTPN QTPVAPHVFA EILQKVVDFF GLANPPAGAF DICKYWSMKR ELTGGTPLTA 540
541 CFENNSLGSL TEEQVQTRID FANDQLEDLY RLKELTTLVK KRTQASNSLS RSRKKVFDIV 600
601 KSPQKYLLKI NVLDIFIKSE QFKALERLVT EPKLLVILEK CKHCDFDTVQ IFKEEIMHFF 660
661 EVLETLPGAS RILQTVSSKA KEQVTNLIGL IEHVDIKKLL SRDFIINDDK IEERPWSGPV 720
721 IMEEEGLSDA EELSAGEHRM LKLILNSG |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
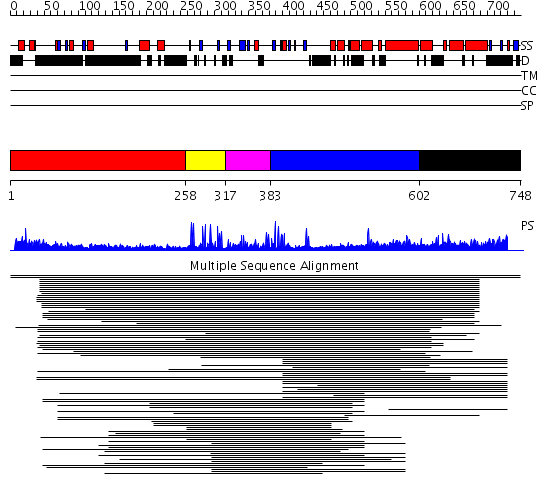
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..257] | 4.257995 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [258..316] | 4.045757 | Williams-Beuren syndrome transcription factor, WSTF | |
| 3 | View Details | [317..382] | 2.300995 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [383..601] | 5.149992 | View MSA. No confident structure predictions are available. | |
| 5 | View Details | [602..748] | 2.39794 | P300/CAF histone acetyltransferase bromodomain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)