













| General Information: |
|
| Name(s) found: |
NCPR_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 691 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPFGIDNTDF TVLAGLVLAV LLYVKRNSIK ELLMSDDGDI TAVSSGNRDI AQVVTENNKN 60
61 YLVLYASQTG TAEDYAKKFS KELVAKFNLN VMCADVENYD FESLNDVPVI VSIFISTYGE 120
121 GDFPDGAVNF EDFICNAEAG ALSNLRYNMF GLGNSTYEFF NGAAKKAEKH LSAAGAIRLG 180
181 KLGEADDGAG TTDEDYMAWK DSILEVLKDE LHLDEQEAKF TSQFQYTVLN EITDSMSLGE 240
241 PSAHYLPSHQ LNRNADGIQL GPFDLSQPYI APIVKSRELF SSNDRNCIHS EFDLSGSNIK 300
301 YSTGDHLAVW PSNPLEKVEQ FLSIFNLDPE TIFDLKPLDP TVKVPFPTPT TIGAAIKHYL 360
361 EITGPVSRQL FSSLIQFAPN ADVKEKLTLL SKDKDQFAVE ITSKYFNIAD ALKYLSDGAK 420
421 WDTVPMQFLV ESVPQMTPRY YSISSSSLSE KQTVHVTSIV ENFPNPELPD APPVVGVTTN 480
481 LLRNIQLAQN NVNIAETNLP VHYDLNGPRK LFANYKLPVH VRRSNFRLPS NPSTPVIMIG 540
541 PGTGVAPFRG FIRERVAFLE SQKKGGNNVS LGKHILFYGS RNTDDFLYQD EWPEYAKKLD 600
601 GSFEMVVAHS RLPNTKKVYV QDKLKDYEDQ VFEMINNGAF IYVCGDAKGM AKGVSTALVG 660
661 ILSRGKSITT DEATELIKML KTSGRYQEDV W |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
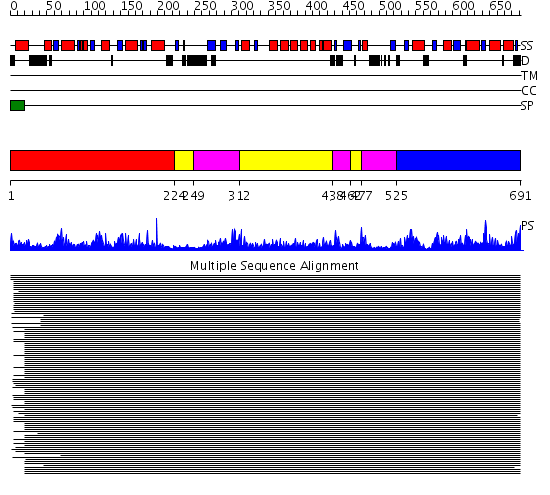
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..223] | 1886.9897 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 2 | View Details | [224..248] [312..437] [462..476] |
1886.9897 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 3 | View Details | [249..311] [438..461] [477..524] |
1886.9897 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain | |
| 4 | View Details | [525..691] | 1886.9897 | NADPH-cytochrome p450 reductase; NADPH-cytochrome p450 reductase, N-terminal domain |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.64 |
Source: Reynolds et al. (2008)