













| General Information: |
|
| Name(s) found: |
NAT1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 854 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSRKRSTKPK PAAKIALKKE NDQFLEALKL YEGKQYKKSL KLLDAILKKD GSHVDSLALK 60
61 GLDLYSVGEK DDAASYVANA IRKIEGASAS PICCHVLGIY MRNTKEYKES IKWFTAALNN 120
121 GSTNKQIYRD LATLQSQIGD FKNALVSRKK YWEAFLGYRA NWTSLAVAQD VNGERQQAIN 180
181 TLSQFEKLAE GKISDSEKYE HSECLMYKND IMYKAASDNQ DKLQNVLKHL NDIEPCVFDK 240
241 FGLLERKATI YMKLGQLKDA SIVYRTLIKR NPDNFKYYKL LEVSLGIQGD NKLKKALYGK 300
301 LEQFYPRCEP PKFIPLTFLQ DKEELSKKLR EYVLPQLERG VPATFSNVKP LYQRRKSKVS 360
361 PLLEKIVLDY LSGLDPTQDP IPFIWTNYYL SQHFLFLKDF PKAQEYIDAA LDHTPTLVEF 420
421 YILKARILKH LGLMDTAAGI LEEGRQLDLQ DRFINCKTVK YFLRANNIDK AVEVASLFTK 480
481 NDDSVNGIKD LHLVEASWFI VEQAEAYYRL YLDRKKKLDD LASLKKEVES DKSEQIANDI 540
541 KENQWLVRKY KGLALKRFNA IPKFYKQFED DQLDFHSYCM RKGTPRAYLE MLEWGKALYT 600
601 KPMYVRAMKE ASKLYFQMHD DRLKRKSDSL DENSDEIQNN GQNSSSQKKK AKKEAAAMNK 660
661 RKETEAKSVA AYPSDQDNDV FGEKLIETST PMEDFATEFY NNYSMQVRED ERDYILDFEF 720
721 NYRIGKLALC FASLNKFAKR FGTTSGLFGS MAIVLLHATR NDTPFDPILK KVVTKSLEKE 780
781 YSENFPLNEI SNNSFDWLNF YQEKFGKNDI NGLLFLYRYR DDVPIGSSNL KEMIISSLSP 840
841 LEPHSQNEIL QYYL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
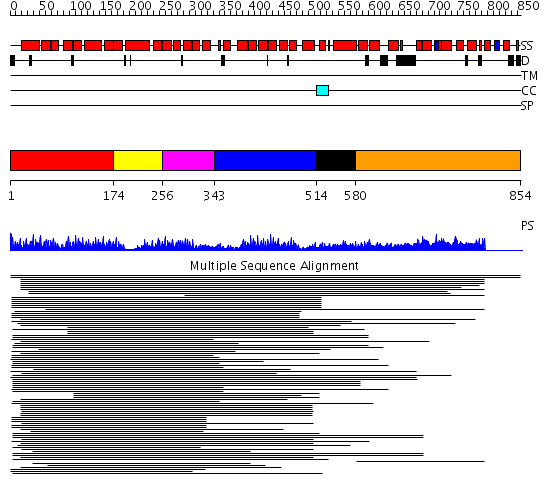
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..173] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 2 | View Details | [174..255] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 3 | View Details | [256..342] | 28.0 | Peroxin pex5 (peroxisomal targeting signal 1 (PTS1) receptor) | |
| 4 | View Details | [343..513] | 20.957997 | View MSA. Confident ab initio structure predictions are available. | |
| 5 | View Details | [514..579] | N/A | No confident structure predictions are available. | |
| 6 | View Details | [580..854] | 1.183954 | View MSA. No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.95 |
Source: Reynolds et al. (2008)