













| General Information: |
|
| Name(s) found: |
NAB6_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1134 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MSNSNSKKPV ANYAYRQQQD YNGMNAMVGN PMMYHPVDFV NGAGQYGPSQ HPAYYTNSPL 60
61 PNIPPTPFDT AYGASLFPSH LLMGSPFVSS PNMQSGYNSA RSSNLKRKAY SRPVSNHNGY 120
121 NGNSNSNQNN TNNGMVTPSN YYRMGRNSFS RNNNSTRNVT HNNNKGCDTR NNSGRRTFAR 180
181 NNIFDDILPE MLLQRPFCIN YKVLPTGDDA YRTRSLLIEN VDHSIDLHSI VKNFVKSNTL 240
241 ESAYLIEGGK SDDSKDVETK NLSILISFLT KGDCLNFYNN ILQRLSEFKT FLKSEALNLK 300
301 FVCLNYDPKC LPTFIESEAL TENAEEADIT NGSTMISASL HHNIANKDAT RSIIIEFKSP 360
361 VEKSDLFKKK LQFLDRSKNK RYILESIDLV NTDVPSNQFP ENYAVLTFLN ISMAIEVLDY 420
421 LKKYSKNLGI SKCFYVSLAP LVVSSARSSV ANIYEGKTST HRLSVPSVTA GNNNDSNNNG 480
481 NNNKSNMSGI TTLNNNSSIG VSVYGHSNMS LTSLSSSVSL NEEIDMLATK LQGVELDGTY 540
541 LEINYRDYQT PTIEEHSTHL SNVKISKTTE NSRQFSQDIP SPLPLNEHMF MNDSNQSNGA 600
601 IIPQQLIATP SPVSPNLQMN QRVLPNPITQ SLEQNFNVSA KVASSMGSDI GNRTIYIGNI 660
661 NPRSKAEDIC NVVRGGILQS IKYIPEKKIC FVTFIEAPSA VQFYANSFID PIVLHGNMLR 720
721 VGWGHYSGPL PKLISLAVTI GASRNVYVSL PEFAFKEKFI HDPQYKKLHE TLSLPDAEQL 780
781 REDFSTYGDI EQINYLSDSH CCWINFMNIS SAISLVEEMN KESTVQNESG EVTLKRATEE 840
841 KFGGRYKGLL INYGKDRCGN INKNLIAGKN SRFYKKVKRP SYNIRLSKLE EKRRQNEIDE 900
901 KEKAFDKPLN LESLGISLDA HKDNGGGETG TANNTGHENE SELEAENENG NETGSFGGLG 960
961 LAVASSDVKR ATSDETDYED IFNKSSGSSD SSSDVEVIMH SPSDPEYALK SQTLRSSSQT1020
1021 VINSKRPVKI EDEEEAVGMS QLNYRSSLRQ APPRAPSTLS YNHSKNNETP MQDIFTNGET1080
1081 ANNRKKKRGS FARHRTIPGS DVMAQYLAQV QHSTFMYAAN ILGASAEDNT HPDE |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
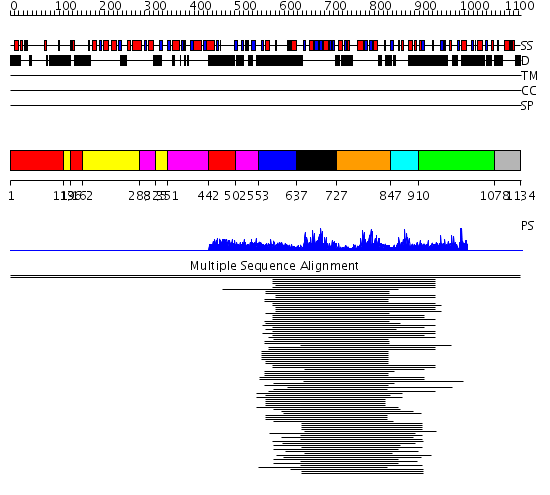
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..118] [136..161] [442..501] |
7.82 | Adenovirus hexon | |
| 2 | View Details | [119..135] [162..287] [325..350] |
7.82 | Adenovirus hexon | |
| 3 | View Details | [288..324] [351..441] [502..552] |
7.82 | Adenovirus hexon | |
| 4 | View Details | [553..636] | 7.97 | Baseplate structural protein gp27 | |
| 5 | View Details | [637..726] | 36.0 | Hu antigen D (Hud) | |
| 6 | View Details | [727..846] | 36.0 | Hu antigen D (Hud) | |
| 7 | View Details | [847..909] | N/A | No confident structure predictions are available. | |
| 8 | View Details | [910..1077] | 1.121988 | View MSA. No confident structure predictions are available. | |
| 9 | View Details | [1078..1134] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||||||||
| 1 | No functions predicted. | ||||||||||||
| 2 |
|
||||||||||||
| 3 | No functions predicted. | ||||||||||||
| 4 | No functions predicted. | ||||||||||||
| 5 | No functions predicted. | ||||||||||||
| 6 | No functions predicted. | ||||||||||||
| 7 | No functions predicted. | ||||||||||||
| 8 | No functions predicted. | ||||||||||||
| 9 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)