













| General Information: |
|
| Name(s) found: |
MUK1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 612 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MARQLFTPPI TNPRFDPNQS IRESYKNTTG GMQFQQNLHE DQNDNERSSC DGDENSTTGE 60
61 RLENNKSPIL TKQEIDEALN TVTNLPPELS KLIDIFIDDL KQPKYVRPLS VLQLSSLFQS 120
121 FYIKFDKASF QHVSSANNNG YYFSGGGSSS FLAAKETLSS GLSGIFGRSR SSSGNSLMRP 180
181 RRSSSLFSNE SISNSTNATQ MLSPEEIKKQ LKINELNNMK IEKYMELCER DVFKKILIVG 240
241 TSVSSPNKMK TFKPHQLQTF KVGNLFRNSV EFTEYNKLLN EKILCLSKLS TMNKINLIKF 300
301 LSLNNGIDPE PKFEEIKDIL YEFTYHSISP CEKIKALLKL HEIMTYSQEM SNDDYLSLLI 360
361 YYIITIVPRD IFLNAEFIRL FRYKKKLVET ESFALTNLEA ALVFVEGLTK NDFSNELQDK 420
421 LTVNESKILE NSISSRVSLP SKTAIMHKNN GNNGSNLGDI VTPTIQRPDV TRSNSYDGFR 480
481 TVFDSSLKNI IGKIRSYTPP HPNNTSNNNL HSSNNLNIPR SSSQLSMELS NRDTTEMSRD 540
541 GSRSTSSSSR SSASLEHGNR EFTGDLTVTA SINGADKKEF QKSWKKYKGY KFEDLTICEL 600
601 RDLFEIYQKM MQ |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
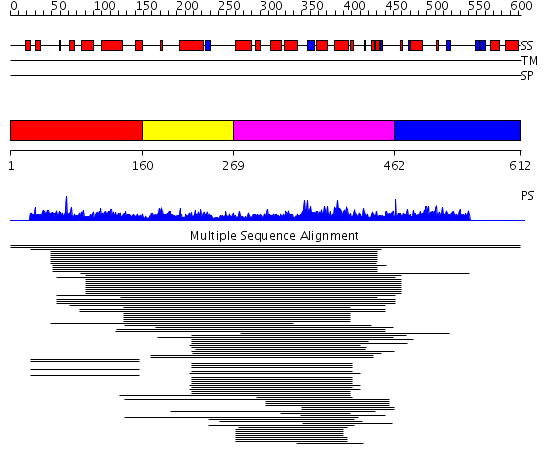
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..159] | 1.020962 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [160..268] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [269..461] | 38.769551 | Vacuolar sorting protein 9 (VPS9) domain No confident structure predictions are available. | |
| 4 | View Details | [462..612] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | ||||||
| 1 | No functions predicted. | ||||||
| 2 |
|
||||||
| 3 | No functions predicted. | ||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.97 |
Source: Reynolds et al. (2008)