













| General Information: |
|
| Name(s) found: |
MPT5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 859 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MINNEPFPSA DSASILTTST SNNSLMSYNH QPQLSINSVQ SLLEPVTPPP LGQMNNKRNH 60
61 QKAHSLDLSG FNQFISSTQS PLALMNNTST SNSANSFSPN PNAASNSTGL SASMANPPAI 120
121 LPLINEFDLE MDGPRRKSSH DFTVVAPSNS GVNTSSLIME TPSSSVTPAA SLRNFSNSNN 180
181 AASKCGVDNS SFGLSSSTSS SMVEISALPL RDLDYIKLAT DQFGCRFLQK KLETPSESNM 240
241 VRDLMYEQIK PFFLDLILDP FGNYLVQKLC DYLTAEQKTL LIQTIYPNVF QISINQYGTR 300
301 SLQKIIDTVD NEVQIDLIIK GFSQEFTSIE QVVTLINDLN GNHVIQKCIF KFSPSKFGFI 360
361 IDAIVEQNNI ITISTHKHGC CVLQKLLSVC TLQQIFKISV KIVQFLPGLI NDQFGNYIIQ 420
421 FLLDIKELDF YLLAELFNRL SNELCQLSCL KFSSNVVEKF IKKLFRIITG FIVNNNGGAS 480
481 QRTAVASDDV INASMNILLT TIDIFTVNLN VLIRDNFGNY ALQTLLDVKN YSPLLAYNKN 540
541 SNAIGQNSSS TLNYGNFCND FSLKIGNLIV LTKELLPSIK TTSYAKKIKL KVKAYAEATG 600
601 IPFTDISPQV TAMSHNNLQT INNENKNPHN KNSHNHNHNH NHNHAHNNNN NNNQKSHTRH 660
661 FSLPANAYHR RSNSSVTNNF SNQYAQDQKI HSPQQIMNFN QNAYPSMGAP SFNSQTNPPL 720
721 VSHNSLQNFD NRQFANLMAH PNSAAPIHSF SSSNITNVNP NVSRGFKQPG FMMNETDKIN 780
781 ANHFSPYSNA NSQNFNESFV PRMQYQTEGA NWDSSLSMKS QHIGQGPYNQ VNMSRNASIS 840
841 NMPAMNTART SDELQFTLP |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
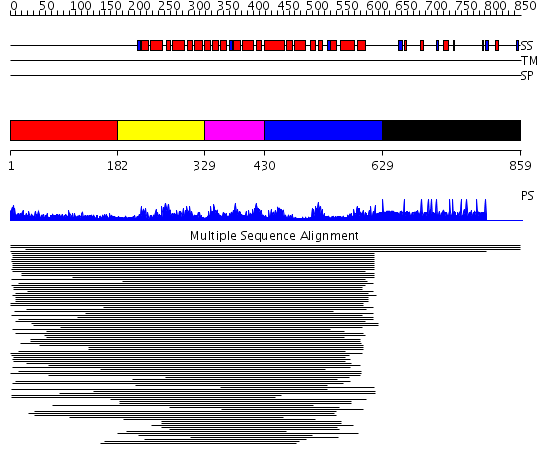
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..181] | 22.163997 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [182..328] | 465.0 | Pumilio 1 | |
| 3 | View Details | [329..429] | 465.0 | Pumilio 1 | |
| 4 | View Details | [430..628] | 465.0 | Pumilio 1 | |
| 5 | View Details | [629..859] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)