













| General Information: |
|
| Name(s) found: |
LCB5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 687 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTLKPSKRRK GRSRHSRKKQ ITSAILTEEG IMIKAKPSSP YTYANRMADK RSRSSIDNIS 60
61 RTSFQSNISR TSFQSNSDNN SIFETASLIS CVTCLSDTDT IDRSETSTTD TSKDDLSANP 120
121 KLHYPSVNGQ LPANTVIPYG RILDARYIEK EPLHYYDANS SPSSPLSSSM SNISEKCDLD 180
181 ELESSQKKER KGNSLSRGSN SSSSLLTSRS PFTKLVEVIF ARPRRHDVVP KRVSLYIDYK 240
241 PHSSSHLKEE DDLVEEILKR SYKNTRRNKS IFVIINPFGG KGKAKKLFMT KAKPLLLASR 300
301 CSIEVVYTKY PGHAIEIARE MDIDKYDTIA CASGDGIPHE VINGLYQRPD HVKAFNNIAI 360
361 TEIPCGSGNA MSVSCHWTNN PSYSTLCLIK SIETRIDLMC CSQPSYAREH PKLSFLSQTY 420
421 GLIAETDINT EFIRWMGPAR FELGVAFNII QKKKYPCEIY VKYAAKSKNE LKNHYLEHKN 480
481 KGSLEFQHIT MNKDNEDCDN YNYENEYETE NEDEDEDADA DDEDSHLISR DLADSSADQI 540
541 KEEDFKIKYP LDEGIPSDWE RLDPNISNNL GIFYTGKMPY VAADTKFFPA ALPSDGTMDM 600
601 VITDARTSLT RMAPILLGLD KGSHVLQPEV LHSKILAYKI IPKLGNGLFS VDGEKFPLEP 660
661 LQVEIMPRLC KTLLRNGRYV DTDFDSM |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
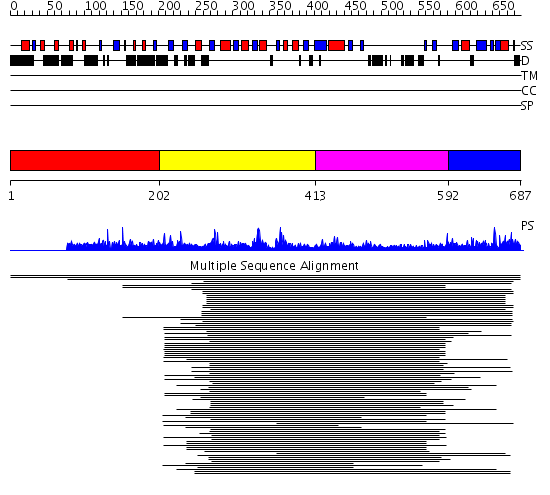
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..201] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [202..412] | 44.60206 | Diacylglycerol kinase catalytic domain (presumed) No confident structure predictions are available. | |
| 3 | View Details | [413..591] | 4.121989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [592..687] | N/A | No confident structure predictions are available. |
Functions predicted (by domain):
| # | Gene Ontology predictions | |||||||||||||||
| 1 | No functions predicted. | |||||||||||||||
| 2 |
|
|||||||||||||||
| 3 | No functions predicted. | |||||||||||||||
| 4 | No functions predicted. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.79 |
Source: Reynolds et al. (2008)