













| General Information: |
|
| Name(s) found: |
KIN4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 800 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MASVPKRHTY GGNVVTDRDR HSLQRNNEIL HPIHKNQRKH ATFGPYIIGS TLGEGEFGKV 60
61 KLGWTKASSS NEVPKQVAIK LIRRDTIKKD ADKEIKIYRE INALKHLTHP NIIYLEEVLQ 120
121 NSKYIGIVLE FVSGGEFYKY IQRKRRLKES SACRLFAQLI SGVNYMHYKG LVHRDLKLEN 180
181 LLLDKHENLV ITDFGFVNEF FEDNELMKTS CGSPCYAAPE LVVSTKAYEA RKADVWSCGV 240
241 ILYAMLAGYL PWDDDHENPT GDDIARLYKY ITQTPLKFPE YITPIPRDLL RRILVPNPRR 300
301 RINLQTIKRH VWLKPHEAFL SIQPNYWDEH LQKERPKPPN KGDVGRHSTY SSSASSYSKS 360
361 RDRNSLIIES TLEQHRMSPQ LATSRPASPT FSTGSKVVLN DTKNDMKESN INGERTSASC 420
421 RYTRDSKGNG QTQIEQVSAR HSSRGNKHTS VAGLVTIPGS PTTARTRNAP SSKLTEHVKD 480
481 SSQTSFTQEE FHRIGNYHVP RSRPRPTSYY PGLSRNTADN SLADIPVNKL GSNGRLTDAK 540
541 DPVPLNAIHD TNKATISNNS IMLLSEGPAA KTSPVDYHYA IGDLNHGDKP ITEVIDKINK 600
601 DLTHKAAENG FPRESIDPES TSTILVTKEP TNSTDEDHVE SQLENVGHSS NKSDASSDKD 660
661 SKKIYEKKRF SFMSLYSSLN GSRSTVESRT SKGNAPPVSS RNPSGQSNRS NIKITQQQPR 720
721 NLSDRVPNPD KKINDNRIRD NAPSYAESEN PGRSVRASVM VSTLREENRS ELSNEGNNVE 780
781 AQTSTARKVL NFFKRRSMRV |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
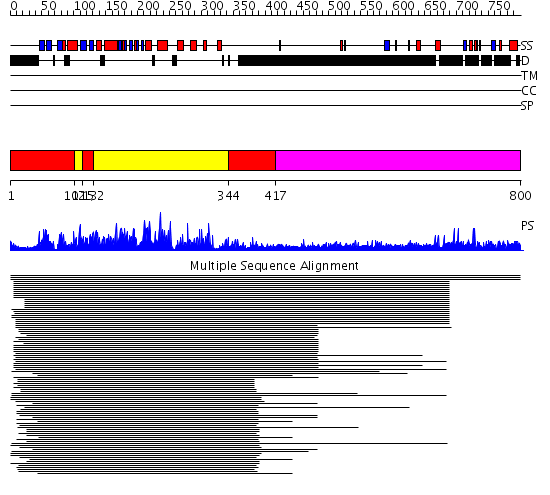
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..101] [115..131] [344..416] |
442.9897 | cAMP-dependent PK, catalytic subunit | |
| 2 | View Details | [102..114] [132..343] |
442.9897 | cAMP-dependent PK, catalytic subunit | |
| 3 | View Details | [417..800] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [558..664] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [665..800] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)