













| General Information: |
|
| Name(s) found: |
KEL1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1164 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MAGFSFAKKF THKKHGKTPS DASISDQSRE ASLSTPPNEK FFTKQETPQK GRQFSQGYHS 60
61 NVNKTSSPPM FARKQVSESR IQPSAVPPQQ RNVSGPSTTL HKQLSKQREY TVWNRIKLQN 120
121 SPFPRYRHVA SAYVTDKNQI YVIGGLHDQS VYGDTWILTA FDNATRFSTT TIDISEATPP 180
181 PRVGHAAVLC GNAFVVFGGD THKVNKEGLM DDDIYLLNIN SYKWTVPAPV GPRPLGRYGH 240
241 KISIIATTQM KTKLYVFGGQ FDDTYFNDLA VYDLSSFRRP DSHWEFLKPR TFTPPPITNF 300
301 TMISYDSKLW VFGGDTLQGL VNDVFMYDPA INDWFIIDTT GEKPPPVQEH ATVVYNDLMC 360
361 VVGGKDEHDA YLNSVYFLNL KSRKWFKLPV FTAGIPQGRS GHSLTLLKND KILIMGGDKF 420
421 DYARVEEYDL HTSDIDMQRG TIVYTLDLAR IKDLCPGVMD VPTDTPTPRN GNLDLATPVT 480
481 PTSHQTKNMN VPISAAPLAS APSPAPKDFS DADRLNREVH NRNVSTEHQN QSHPVNSESH 540
541 LIAEPNILTP YVPSESSQTP VMKITSNKPF DTPTIQKEPD LSETMDPTVG NQRIPSSIYG 600
601 DNLTPANQIK NNSPILETLP SNEIKTPQNG NIEEIKHLPD ADEKIDSTTT FDQEINGDKL 660
661 GTSSMSKVEE DGNVADEDDE IGVAQMASSP SKDQFKIKHY NESSELSQNN TEIDKLSEPV 720
721 DITIKKSDTA GHDSANHVID ASDEKNVSPM GDVPTDTKNE EASVPINRDA TTEVVDRALF 780
781 EKLRSELQSL KELTHEKALE AGAHIKELET ELWQLKSQKN SGTTKEIDEL DSVRLQSKCE 840
841 ILEADNHSLE DKVNELEELV NSKFLDIENL NEVIQFQNEK IKSLELEPNY KEKLEELQIE 900
901 HENLSRENER LKNESKQHNE DIINNVANYS SQLGSLISHW KENRANSSFL ESSSSLISVS 960
961 DENGEKTVGE PYGDQSRHHR VVINKLTNRL DDLLERSQEL TISKEKLSSE YHALKMEHSS1020
1021 LSQDVLVKEN EIKKIQNDYK ESISSMDSAS KALMVSQREL EKYKSLNKKL IDELDELKFK1080
1081 NGVCSENFEN GLRSTEESSN NVKNSNSIRE NQFNIKINDL KAELFITNQE RDDLKSEVLE1140
1141 LKKRLLNLEN NTKQVNEDAD SDLL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
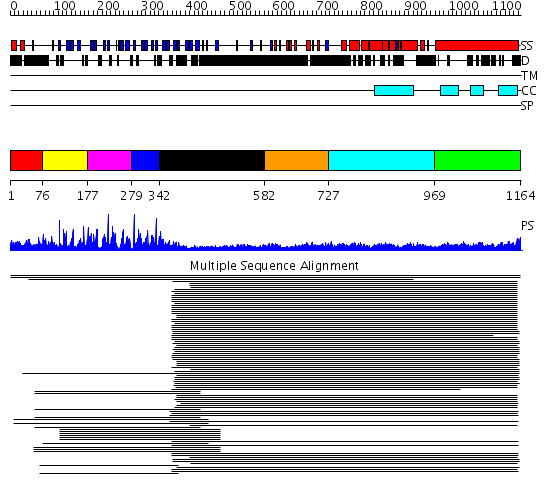
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..75] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [76..176] | 2.260997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [177..278] | 2.481486 | Kelch motif No confident structure predictions are available. | |
| 4 | View Details | [279..341] | 5.244125 | Kelch motif No confident structure predictions are available. | |
| 5 | View Details | [342..581] | 4.30103 | Interferon-induced guanylate-binding protein 1 (GBP1), C-terminal domain; Interferon-induced guanylate-binding protein 1 (GBP1), N-terminal domain | |
| 6 | View Details | [582..726] | 7.154902 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 7 | View Details | [727..968] | 7.154902 | Colicin Ia; Colicin Ia, N-terminal domain | |
| 8 | View Details | [969..1164] | 23.69897 | Heavy meromyosin subfragment |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)