













| General Information: |
|
| Name(s) found: |
KCS1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1050 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDTSHEIHDK IPDTLREQQQ HLRQKESEGC ITTLKDLNVP ETKKLSSVLH GRKASTYLRI 60
61 FRDDECLADN NNGVDSNNGG SVTCADKITR SEATPKSVPE GLQVSEKKNN PDTLSSSLSS 120
121 FILSNHEEPA IKPNKHVAHR NNITETGQGS GEDIAKQQSH QPQVLHHQTS LKPIQNVDEG 180
181 CISPKSTYQE SLHGISEDLT LKPVSSATYY PHKSKADSGY EEKDKMENDI DTIQPATINC 240
241 ASGIATLPSS YNRHTFKVKT YSTLSQSLRQ ENVNNRSNEK KPQQFVPHSE SIKEKPNTFE 300
301 QDKEGEQADE EEDEGDNEHR EYPLAVELKP FTNRVGGHTA IFRFSKRAVC KALVNRENRW 360
361 YENIELCHKE LLQFMPRYIG VLNVRQHFQS KDDFLSDLDQ ENNGKNDTSN ENKDIEVNHN 420
421 NNDDIALNTE PTGTPLTHIH SFPLEHSSRQ VLEKEHPEIE SVHPHVKRSL SSSNQPSLLP 480
481 EVVLNDNRHI IPESLWYKYS DSPNSAPNDS YFSSSSSHNS CSFGERGNTN KLKRRDSGST 540
541 MINTELKNLV IREVFAPKCF RRKRNSNTTT MGNHNARLGS SPSFLTQKSR ASSHDASNTS 600
601 MKTLGDSSSQ ASLQMDDSKV NPNLQDPFLK KSLHEKISNA LDGSHSVMDL KQFHKNEQIK 660
661 HKNSFCNSLS PILTATNSRD DGEFATSPNY ISNAQDGVFD MDEDTGNETI NMDNHGCHLD 720
721 SGKNMIIKSL AYNVSNDYSH HDIESITFEE TSHTIVSKFI LLEDLTRNMN KPCALDLKMG 780
781 TRQYGVDAKR AKQLSQRAKC LKTTSRRLGV RICGLKVWNK DYYITRDKYF GRRVKVGWQF 840
841 ARVLARFLYD GKTIESLIRQ IPRLIKQLDT LYSEIFNLKG YRLYGASLLL MYDGDANKSN 900
901 SKRKKAANVK VNLIDFARCV TKEDAMECMD KFRIPPKSPN IEDKGFLRGV KSLRFYLLLI 960
961 WNYLTSDMPL IFDEVEMNDM ISEEADSNSF TSATGSKINF NSKWDWLDEF DKEDEEMYND1020
1021 PNSKLRQKWR KYELIFDAEP RYNDDAQVSD |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
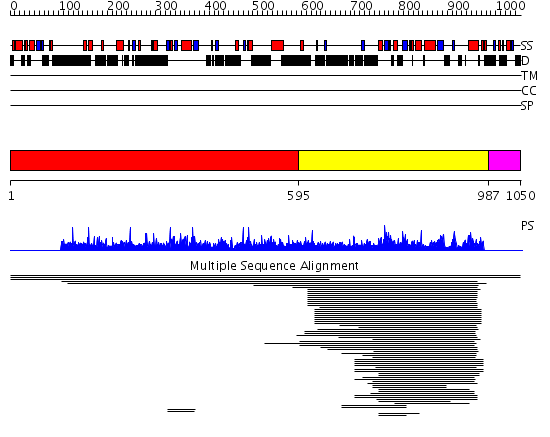
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..594] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [595..986] | 23.040959 | Inositol polyphosphate kinase No confident structure predictions are available. | |
| 3 | View Details | [987..1050] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [965..1050] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)