













| General Information: |
|
| Name(s) found: |
KCC4_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1037 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MTVANTETHS AAKPSSTIGP WKLGETLGFG STGKVQLAQH ERTGHRTAVK VISKSIFNNN 60
61 GNHSNDDSVL PYNIEREIVI MKLLSHPNVL SLYDVWETNN NLYLILEYAE KGELFNLLVD 120
121 HGPLPEREAI NCFRQIIIGI SYCHALGIVH RDLKPENLLL DSFYNIKIAD FGMAALQTDA 180
181 DLLETSCGSP HYAAPEIVSG LPYEGFASDV WSCGVILFAL LTGRLPFDEE NGNVRDLLLK 240
241 VQKGQFEMPN DTEISRDAQD LIGKILVVDP RQRIKIRDIL SHPLLKKYQT IKDSKSIKDL 300
301 PRENTYLYPL ADSNNHTSAS IDDSILQNLV VLWHGRHADD IVSKLKENGT NKEKILYALL 360
361 YRFKLDSVRG SNKKNRNKIK KTKKNKRSST LSSSSSLLLN NRSIQSTPRR RTSKRHSREF 420
421 SSSRKRSSFL LSSNPTDSSP IPLRSSKRIT HINVASANTQ ATPSGVPNPH KRNSKKRSSK 480
481 RLSYMPNTKR SSLTSKSLSN FTNLIDDDDW EYIEKDAKRT SSNFATLIDE IFEPEKFELA 540
541 KREKAELQRK VQEAKRQSVN AQKINEDEFG SEVSDGMKEL KKINDKVSSP LINYEFSQQE 600
601 LLQDIDTLLT NRYQLSSYTR PISRLDPGLT PVTETLPNNL KEKTALLQDT EKKIIETIRR 660
661 SKFLGSLLNV RGGLSPGKSE LAPIEESPIV STTPLIYNDR MEPRRISDVE VPHFTRKSKH 720
721 FTTANNRRSV LSLYAKDSIK DLNEFLIKED PDLPPQGSTD NESRSEDPEI AESITDSRNI 780
781 QYDEDDSKDG DNVNNDNILS DFPQGVGISQ EYDMKDKNPN QSPISKSAEP TLVVKLPSLS 840
841 SFQGKNASGL GLYQREPSKV TLPSLTSNNS SVGENIEDGA EKGTESEKIA ASLSDDDLKE 900
901 DNDKKDNDTV NAPTTVKKPP NSVLLKKFSK GKILELEIHA KIPEKRLYEG LHKLLEGWKQ 960
961 YGLKNLVFNI TNMIITGKLV NDSILFLRST LFEIMVLPNG DGRSLIKFNK KTGSTKTLTK1020
1021 LATEIQIILQ KEGVLDK |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
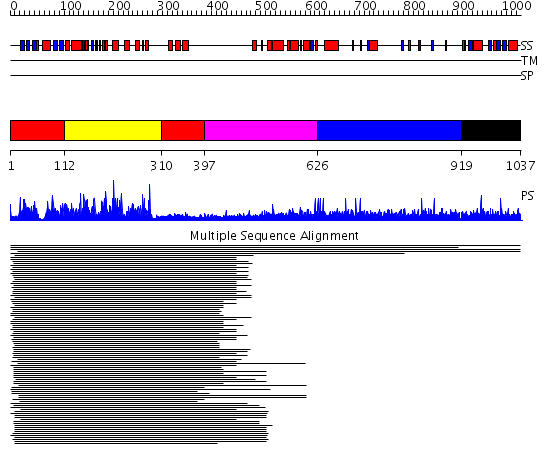
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..111] [310..396] |
557.9897 | Calmodulin-dependent protein kinase | |
| 2 | View Details | [112..309] | 557.9897 | Calmodulin-dependent protein kinase | |
| 3 | View Details | [397..625] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [626..918] | 7.24 | Microbial transglutaminase | |
| 5 | View Details | [919..1037] | N/A | Confident ab initio structure predictions are available. | |
| 6 | View Details | [921..1037] | N/A | Confident ab initio structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)