













| General Information: |
|
| Name(s) found: |
IMB5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1004 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MDINELIIGA QSADKHTREV AETQLLQWCD SDASQVFKAL ANVALQHEAS LESRQFALLS 60
61 LRKLITMYWS PGFESYRSTS NVEIDVKDFI REVLLKLCLN DNENTKIKNG ASYCIVQISA 120
121 VDFPDQWPQL LTVIYDAISH QHSLNAMSLL NEIYDDVVSE EMFFEGGIGL ATMEIVFKVL 180
181 NTETSTLIAK IAALKLLKAC LLQMSSHNEY DEASRKSFVS QCLATSLQIL GQLLTLNFGN 240
241 VDVISQLKFK SIIYENLVFI KNDFSRKHFS SELQKQFKIM AIQDLENVTH INANVETTES 300
301 EPLLETVHDC SIYIVEFLTS VCTLQFSVEE MNKIITSLTI LCQLSSETRE IWTSDFNTFV 360
361 SKETGLAASY NVRDQANEFF TSLPNPQLSL IFKVVSNDIE HSTCNYSTLE SLLYLLQCIL 420
421 LNDDEITGEN IDQSLQILIK TLENILVSQE IPELILARAI LTIPRVLDKF IDALPDIKPL 480
481 TSAFLAKSLN LALKSDKELI KSATLIAFTY YCYFAELDSV LGPEVCSETQ EKVIRIINQV 540
541 SSDAEEDTNG ALMEVLSQVI SYNPKEPHSR KEILQAEFHL VFTISSEDPA NVQVVVQSQE 600
601 CLEKLLDNIN MDNYKNYIEL CLPSFINVLD SNNANNYRYS PLLSLVLEFI TVFLKKKPND 660
661 GFLPDEINQY LFEPLAKVLA FSTEDETLQL ATEAFSYLIF NTDTRAMEPR LMDIMKVLER 720
721 LLSLEVSDSA AMNVGPLVVA IFTRFSKEIQ PLIGRILEAV VVRLIKTQNI STEQNLLSVL 780
781 CFLTCNDPKQ TVDFLSSFQI DNTDALTLVM RKWIEAFEVI RGEKRIKENI VALSNLFFLN 840
841 DKRLQKVVVN GNLIPYEGDL IITRSMAKKM PDRYVQVPLY TKIIKLFVSE LSFQSKQPNP 900
901 EQLITSDIKQ EVVNANKDDD NDDWEDVDDV LDYDKLKEYI DDDVDEEADD DSDDITGLMD 960
961 VKESVVQLLV RFFKEVASKD VSGFHCIYET LSDSERKVLS EALL |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
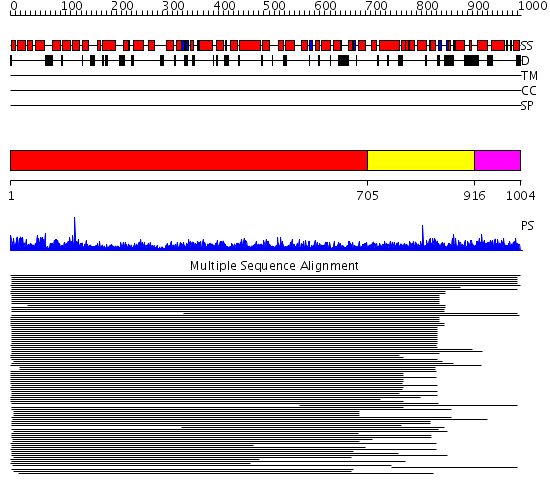
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..704] | 13.126979 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [705..915] | 2.087982 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [916..1004] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.98 |
Source: Reynolds et al. (2008)