













| General Information: |
|
| Name(s) found: |
HUL5_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 910 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MLNFTGQTRR RNVNLGNRTR NSKKDLLEKA KRERERRAQD KLKEDASKTI QKSIRRHFSN 60
61 VRLFKNTFTS SQLVHMIPAY GGKLIYYISQ YDLQQLLKLS HNFLSSYPNS LGNRQLLSLL 120
121 KLYQDDALVA ETLSDLNMDC PTVDEFLDSL SVYLCRASFL SYSSASKLAD VIEAWEVMHS 180
181 SASISIFSIS IGSYEKRPFA LQFYCILAER NLLPQLINTN PILWDNMAKT YSHCSKGGQK 240
241 NIAKLLIPNF NNHIAPSVLR SDNDYVLKFY EKAFIDEVIA TTANYVSDED HVKNLMCYIA 300
301 SSPNQSCKNS VLITLLSNKD FVRRLSWEFF HTKFNASKTE AHPLFSVLAQ LIDMHLLIST 360
361 DRELLDYNSV IPIEELKKFT STLKDFTFRQ YWELPKSERN PMLKEAVPLL SKVYERDSRL 420
421 HFLSTENNPT YWENSEKQFL NLRFYEELQE YEDLYREHLE EESDEDMEKE IDLDKERPPL 480
481 KSLLLNKMKK RLKSSLRFRK LEILLELPFF IPFEERVDLF YMFIALDKKR LSLDDDHNLI 540
541 NMFTPWASTG MRKQSAIISR DNVLEDAFNA FNSIGERFKA SLDVTFINEF GEEAGIDGGG 600
601 ITKEFLTTVS DEGFKDPKHE LFRTNDRYEL YPSVVYDATK LKYIWFLGKV VGKCLYEHVL 660
661 IDVSFADFFL KKLLNYSNGF LSSFSDLGSY DSVLYNNLIK LLNMTTDEIK SLDLTFEIDE 720
721 PESSAKVVDL IPNGSKTYVT KDNVLLYVTK VTDYKLNKRC FKPVSAFHGG LSVIIAPHWM 780
781 EMFNSIELQM LISGERDNID LDDLKSNTEY GGYKEEDQTI VDFWEVLNEF KFEEKLNFLK 840
841 FVTSVPQAPL QGFKALDPKF GIRNAGTEKY RLPTASTCVN LLKLPDYRNK TILREKLLYA 900
901 INSGARFDLS |
SHOWING SINGLE HITS. [ Hide Single Hits ]
New Feature: Upload Your Own Microscopy Data
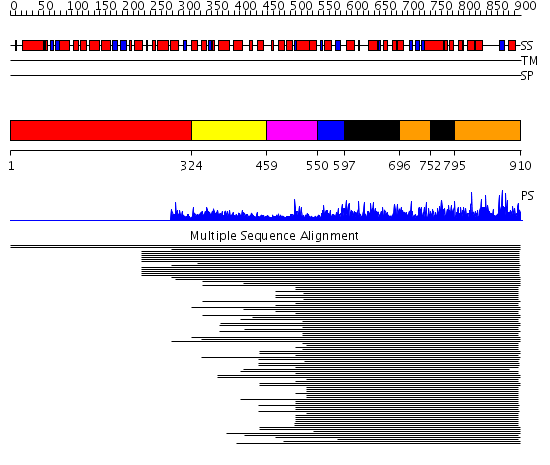
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..323] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [324..458] | 1.04399 | View MSA. Confident ab initio structure predictions are available. | |
| 3 | View Details | [459..549] | N/A | No confident structure predictions are available. | |
| 4 | View Details | [550..596] | 504.228787 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 5 | View Details | [597..695] [752..794] |
504.228787 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) | |
| 6 | View Details | [696..751] [795..910] |
504.228787 | Ubiquitin-protein ligase E3a, Hect catalytic domain (E6ap) |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.93 |
Source: Reynolds et al. (2008)