













| General Information: |
|
| Name(s) found: |
HBT1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 1046 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MNMNESISKD GQGEEEQNNF SFGGKPGSYD SNSDSAQRKK SFSTTKPTEY NLPKEQPEST 60
61 SKNLETKAKN ILLPWRKKHN KDSETPHEDT EADANRRANV TSDVNPVSAD TKSSSGPNAT 120
121 ITTHGYSYVK TTTPAATSEQ SKVKTSPPTS HEHSNIKASP TAHRHSKGDA GHPSIATTHN 180
181 HSTSKAATSP VTHTHGHSSA TTSPVTHTHG HASVKTTSPT NTHEHSKANT GPSATATTHG 240
241 HINVKTTHPV SHGHSGSSTG PKSTAAAQDH SSTKTNPSVT HGHTSVKDNS SATKGYSNTD 300
301 SNSDRDVIPG SFRGMTGTDV NPVDPSVYTS TGPKSNVSSG MNAVDPSVYT DTSSKSADRR 360
361 KYSGNTATGP PQDTIKEIAQ NVKMDESEQT GLKNDQVSGS DAIQQQTMEP EPKAAVGTSG 420
421 FVSQQPSYHD SNKNIQHPEK NKVDNKNISE RAAEKFNIER DDILESADDY QQKNIKSKTD 480
481 SNWGPIEYSS SAGKNKNLQD VVIPSSMKEK FDSGTSGSQN MPKAGTELGH MKYNDNGRDN 540
541 LQYVAGSQAG SQNTNNNIDM SPRHEAEWSG LSNDATTRNN VVSPAMKDED MNEDSTKPHQ 600
601 YGLDYLDDVE DYHENDIDDY SNAKKNDLYS KKAYQGKPSD YNYEQREKIP GTFEPDTLSK 660
661 SVQKQDEDPL SPRQTTNRAG METARDESLG NYEYSNTSGN KKLSDLSKNK SGPTPTRSNF 720
721 IDQIEPRRAK TTQDIASDAK DFTNNPETGT TGNVDTTGRM GAKSKTFSSN PFDDSKNTDT 780
781 HLENANVAAF DNSRSGDTTY SKSGDAETAA YDNIKNADPT YAKSQDITGM THDQEPSSEQ 840
841 KASYGSGGNS QNQEYSSDDN IDVNKNAKVL EEDAPGYKRE VDLKNKRRTD LGGADASNAY 900
901 AAEVGNFPSL IDPHVPTYGF KDTNTSSSQK PSEGTYPETT SYSIHNETTS QGRKVSVGSM 960
961 GSGKSKHHHN HHRHSRQNSS KGSDYDYNNS THSAEHTPRH HQYGSDEGEQ DYHDDEQGEE1020
1021 QAGKQSFMGR VRKSISGGTF GFRSEI |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
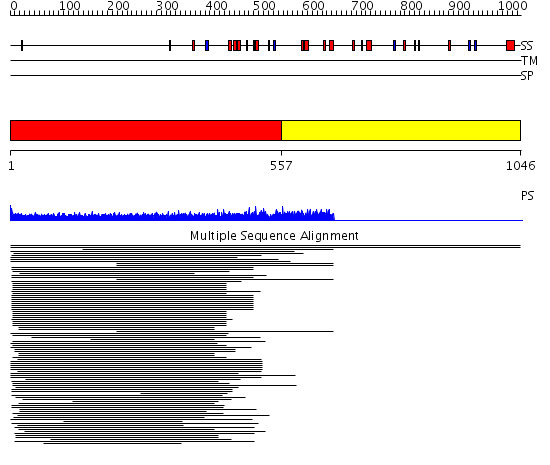
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..556] | 56.290998 | View MSA. No confident structure predictions are available. | |
| 2 | View Details | [557..1046] | N/A | No confident structure predictions are available. | |
| 3 | View Details | [396..569] | 1.130989 | View MSA. No confident structure predictions are available. | |
| 4 | View Details | [570..771] | N/A | No confident structure predictions are available. | |
| 5 | View Details | [772..1046] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.99 |
Source: Reynolds et al. (2008)