













| General Information: |
|
| Name(s) found: |
GPB1_YEAST
[Swiss-Prot]
|
| Description(s) found: |
|
| Organism: | Saccharomyces cerevisiae S288c |
| Length: | 897 amino acids |
Gene Ontology: |
|
| Cellular Component: | NONE FOUND |
| Biological Process: | NONE FOUND |
| Molecular Function: | NONE FOUND |
| Sequence: |
|
| Sequence: [PDR BLAST] [ProtParam] |
1 11 21 31 41 51
| | | | | |
1 MPQASTFGSH SLEAHPLHIQ PAVHIKLSKE ERSHYREQYD SLKYISNYVS VFDQALSDNI 60
61 DSRIRKENEA LLKKYYESRK PFTFTSFRQG SVISSSDSST GFTERTKTYC FLNDFVSNCV 120
121 NEVDPYTLKM TVRNRNTALN MENLDDERKS KDDIYDFEDN TDDECNAKCH GAFHYSSERL 180
181 EILRSRSTIS YFKYYKKLLT VDLRDSDVLK RHNLWMPMIT RRFRFLLVSS SKPEDVRLTT 240
241 PIPTFSESDL DIFKNKTCPL FINGTDCVPR SYDTFSGSSV IASIFSEYKL PSLSYHCSVE 300
301 LNDQLFIVGG LMACHRYDEE APDLKDFYVD GIKNLPPPLI PELINNPSMI PNPHLYCFSL 360
361 TSSRLTRPDI SGYIPPPLVC TQGCKLTERH IFLYGGFEIK SETQVDDKGR YFIRKRAFLN 420
421 NTGYILDTVT FNFSKIELVA PPYQFAIYNN FSPRFGHMQA SISNSNNNVS NENTTTSAKG 480
481 RRSISPYRQG NGDHKIDDLV GSPGSTDYLE DDAIPPVTNP RSTDSLSSKH CSTATHICSS 540
541 VNTILIFGGY SQTGDDKYEA MNDMWKINIP VVSRGKRNYY KFADTVTATK IPIIDDPELW 600
601 PSRRAFSACC VPDYFTKDVE PIETRLLRNL KNDFSIDLEI RPGNKPSQPL FPNIPHSRKE 660
661 KKSGRDSMHI SNSNNSTSED TSSKSTRNTT SSPPTSPKHT PPLNPSKKCA SIGRTIAFHG 720
721 GSDGYDVCSD MWWFDFDSET WTKIDLYAKT QEESDGLVPI NLCMVGHSMT TVGHKVVLIG 780
781 GLRQGDVDRI YRDETLPEEV ISGVPLGSGV INVVDLNTQC LQGCKLIRND GDTKESVIMD 840
841 PHVGTPHQVL AVAGTIELVK GTMTLIGGVV AGREDISSLY LRGAVLQFIL PSMNLAN |
NOT SHOWING SINGLE HITS. [ Show Single Hits ]
New Feature: Upload Your Own Microscopy Data
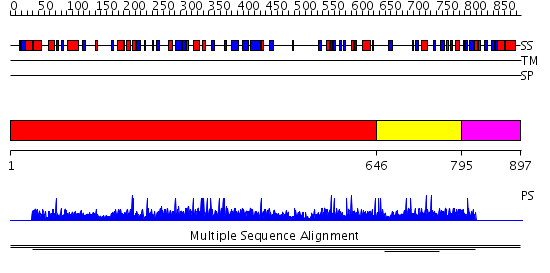
Domains predicted:
| # | Region(s) | Method | Confidence | Match Description | |
| 1 | View Details | [1..645] | N/A | No confident structure predictions are available. | |
| 2 | View Details | [646..794] | 1.003997 | View MSA. No confident structure predictions are available. | |
| 3 | View Details | [795..897] | N/A | No confident structure predictions are available. |
| Protein predicted to be: | GLOBULAR (No transmembrane regions or signal peptide) |
| Confidence of classification: | 0.96 |
Source: Reynolds et al. (2008)